Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105374 |
| Name | oriT_p30177-1 |
| Organism | Salmonella enterica subsp. enterica serovar Kentucky strain CVM 30177 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP051347 (44645..44997 [-], 353 nt) |
| oriT length | 353 nt |
| IRs (inverted repeats) | 256..263, 272..279 (ACCGCTAG..CTAGCGGT) 192..198, 206..212 (TATAAAA..TTTTATA) 40..47, 50..57 (GCAAAAAC..GTTTTTGC) 4..11, 16..23 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 353 nt
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGTGTTATGAAAAATTAGTTTCTCTTACTCTCTTTATGATATTTAAAAAAGCGGTGTCGGCGCGGCTACAACAACGCGCCGACACCGTTTTGTAGGGGTGGTACTGACTATTTTTATAAAAAACATTATTTTATATTAGGGGGGGCTGCTAGCGGCGCGGTGTGTTTTTTTATAGGATACCGCTAGGGGCGCTGCTAGCGGTGCGTCCCTGTTTGCATTATGAATTTTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGTAATCTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file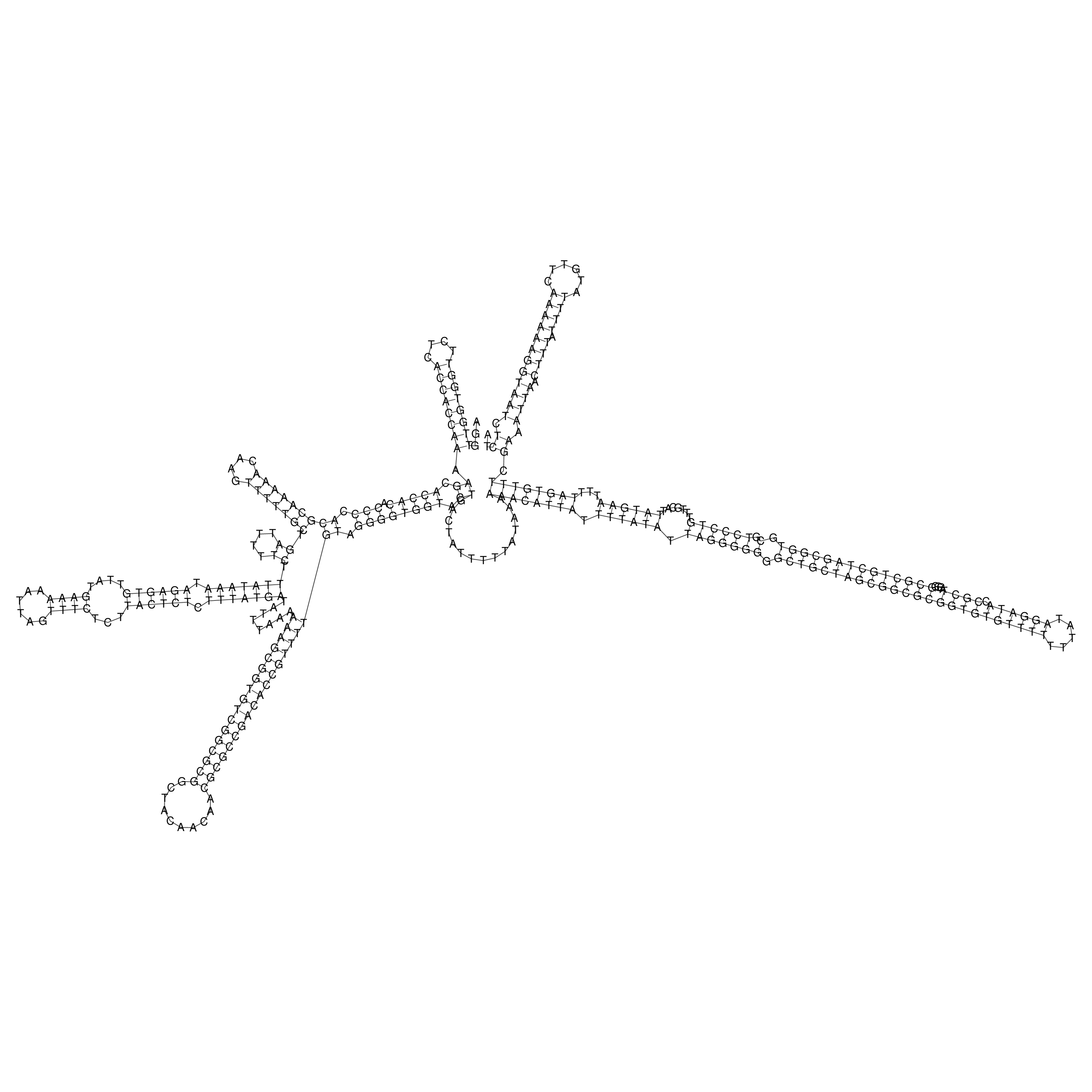
Relaxase
| ID | 3668 | GenBank | WP_000986988 |
| Name | traI_HFT18_RS22875_p30177-1 |
UniProt ID | A0A5V2AWD5 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191867.46 Da Isoelectric Point: 5.6862
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNRHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRIMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFVASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDVMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLESAISHQKSALHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGETFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDNVYHRIAGISRDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQEQAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRADITEREAALPESVLRESQREQEAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1656 | GenBank | WP_001369361 |
| Name | WP_001369361_p30177-1 |
UniProt ID | B7LI64 |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15409.70 Da Isoelectric Point: 9.9240
MLLKRFGTRSATGKMVKLKLPVDVESLLIEASNRSGRSRSFEAVIRLKDHLHRYPKFNRAGNIYGKSLVK
YLTMRLDDETNQLLIAAKNRSGWCKTDEAADRVIDHLIKFPDFYNSEIFREADKEEDITFNTL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B7LI64 |
| ID | 1657 | GenBank | WP_001151524 |
| Name | WP_001151524_p30177-1 |
UniProt ID | A0A5W2DT22 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14517.48 Da Isoelectric Point: 5.0516
MAKVNLYISNDAYEKINAIIEKRRQEGAREKDVSFSATASMLLELGLRVHEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5W2DT22 |
T4CP
| ID | 3643 | GenBank | WP_000009364 |
| Name | traD_HFT18_RS22880_p30177-1 |
UniProt ID | _ |
| Length | 735 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 735 a.a. Molecular weight: 83567.22 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQ
QENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3644 | GenBank | WP_001064251 |
| Name | traC_HFT18_RS22960_p30177-1 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99173.85 Da Isoelectric Point: 5.9407
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASITTQTVDAQAFIDIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRESGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAESPTIRSRLDEMIVLLDQYT
ANGTYGQYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGWR
LLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLHVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEGL
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 19760..45568
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HFT18_RS22880 (HFT18_22870) | 19760..21967 | - | 2208 | WP_000009364 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HFT18_RS22885 (HFT18_22875) | 22219..22950 | - | 732 | WP_001447147 | conjugal transfer complement resistance protein TraT | - |
| HFT18_RS22890 (HFT18_22880) | 22982..23485 | - | 504 | WP_000605510 | hypothetical protein | - |
| HFT18_RS22895 (HFT18_22885) | 23500..26322 | - | 2823 | WP_001007012 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HFT18_RS22900 (HFT18_22890) | 26319..27695 | - | 1377 | WP_000982833 | conjugal transfer pilus assembly protein TraH | traH |
| HFT18_RS22905 (HFT18_22895) | 27692..28054 | - | 363 | WP_071779407 | P-type conjugative transfer protein TrbJ | - |
| HFT18_RS22910 (HFT18_22900) | 27984..28529 | - | 546 | WP_000052618 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HFT18_RS22915 (HFT18_22905) | 28516..28800 | - | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HFT18_RS22920 (HFT18_22910) | 28919..29266 | - | 348 | WP_001287676 | conjugal transfer protein TrbA | - |
| HFT18_RS22925 (HFT18_22915) | 29282..30025 | - | 744 | WP_001030389 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HFT18_RS22930 (HFT18_22920) | 30018..30275 | - | 258 | WP_000864325 | conjugal transfer protein TrbE | - |
| HFT18_RS22935 (HFT18_22925) | 30302..32110 | - | 1809 | WP_000821846 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HFT18_RS22940 (HFT18_22930) | 32107..32745 | - | 639 | WP_022630459 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HFT18_RS22945 (HFT18_22935) | 32754..33746 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| HFT18_RS22950 (HFT18_22940) | 33743..34375 | - | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HFT18_RS22955 (HFT18_22945) | 34372..34758 | - | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| HFT18_RS22960 (HFT18_22950) | 34755..37382 | - | 2628 | WP_001064251 | type IV secretion system protein TraC | virb4 |
| HFT18_RS22965 (HFT18_22955) | 37542..37763 | - | 222 | WP_001278689 | conjugal transfer protein TraR | - |
| HFT18_RS22970 (HFT18_22960) | 37898..38413 | - | 516 | WP_000809841 | type IV conjugative transfer system lipoprotein TraV | traV |
| HFT18_RS22975 (HFT18_22965) | 38410..38661 | - | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| HFT18_RS22980 (HFT18_22970) | 38673..38870 | - | 198 | WP_001324648 | conjugal transfer protein TrbD | - |
| HFT18_RS22985 (HFT18_22975) | 38857..39447 | - | 591 | WP_000002796 | conjugal transfer pilus-stabilizing protein TraP | - |
| HFT18_RS22990 (HFT18_22980) | 39437..40864 | - | 1428 | WP_000146685 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HFT18_RS22995 (HFT18_22985) | 40864..41592 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| HFT18_RS23000 (HFT18_22990) | 41579..42145 | - | 567 | WP_000399785 | type IV conjugative transfer system protein TraE | traE |
| HFT18_RS23005 (HFT18_22995) | 42167..42478 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HFT18_RS23010 (HFT18_23000) | 42493..42858 | - | 366 | WP_000994779 | type IV conjugative transfer system pilin TraA | - |
| HFT18_RS23015 (HFT18_23005) | 42891..43286 | - | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| HFT18_RS23020 (HFT18_23010) | 43385..44074 | - | 690 | WP_000283385 | conjugal transfer transcriptional regulator TraJ | - |
| HFT18_RS23025 (HFT18_23015) | 44261..44644 | - | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HFT18_RS23030 (HFT18_23020) | 45059..45568 | + | 510 | WP_000759163 | transglycosylase SLT domain-containing protein | virB1 |
| HFT18_RS23035 (HFT18_23025) | 45864..46685 | - | 822 | WP_001234468 | DUF932 domain-containing protein | - |
| HFT18_RS23040 (HFT18_23030) | 46804..47091 | - | 288 | WP_000107520 | hypothetical protein | - |
| HFT18_RS23045 (HFT18_23040) | 47392..47689 | + | 298 | Protein_52 | hypothetical protein | - |
| HFT18_RS23050 (HFT18_23045) | 47990..48112 | - | 123 | WP_223195199 | Hok/Gef family protein | - |
| HFT18_RS24080 | 48057..48209 | - | 153 | Protein_54 | DUF5431 family protein | - |
| HFT18_RS23055 (HFT18_23050) | 48360..49146 | - | 787 | Protein_55 | plasmid SOS inhibition protein A | - |
| HFT18_RS23060 (HFT18_23055) | 49143..49577 | - | 435 | WP_000845922 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 5812 | GenBank | NZ_CP051347 |
| Plasmid name | p30177-1 | Incompatibility group | IncFIB |
| Plasmid size | 146811 bp | Coordinate of oriT [Strand] | 44645..44997 [-] |
| Host baterium | Salmonella enterica subsp. enterica serovar Kentucky strain CVM 30177 |
Cargo genes
| Drug resistance gene | tet(B), sitABCD, aph(6)-Id, aph(3'')-Ib |
| Virulence gene | iutA, iucC, iucB, iucA, iroB, iroC, iroD, iroE, iroN |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |