Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105341 |
| Name | oriT_pBs162S2c |
| Organism | Bradyrhizobium septentrionale strain 162S2 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP088291 (9113..9168 [-], 56 nt) |
| oriT length | 56 nt |
| IRs (inverted repeats) | 23..29, 34..40 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 56 nt
>oriT_pBs162S2c
CCCCGGCAGGCGGGAAAACAGGACGTCGCGGATGCGACGTATTACTGCGCCCTTGG
CCCCGGCAGGCGGGAAAACAGGACGTCGCGGATGCGACGTATTACTGCGCCCTTGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file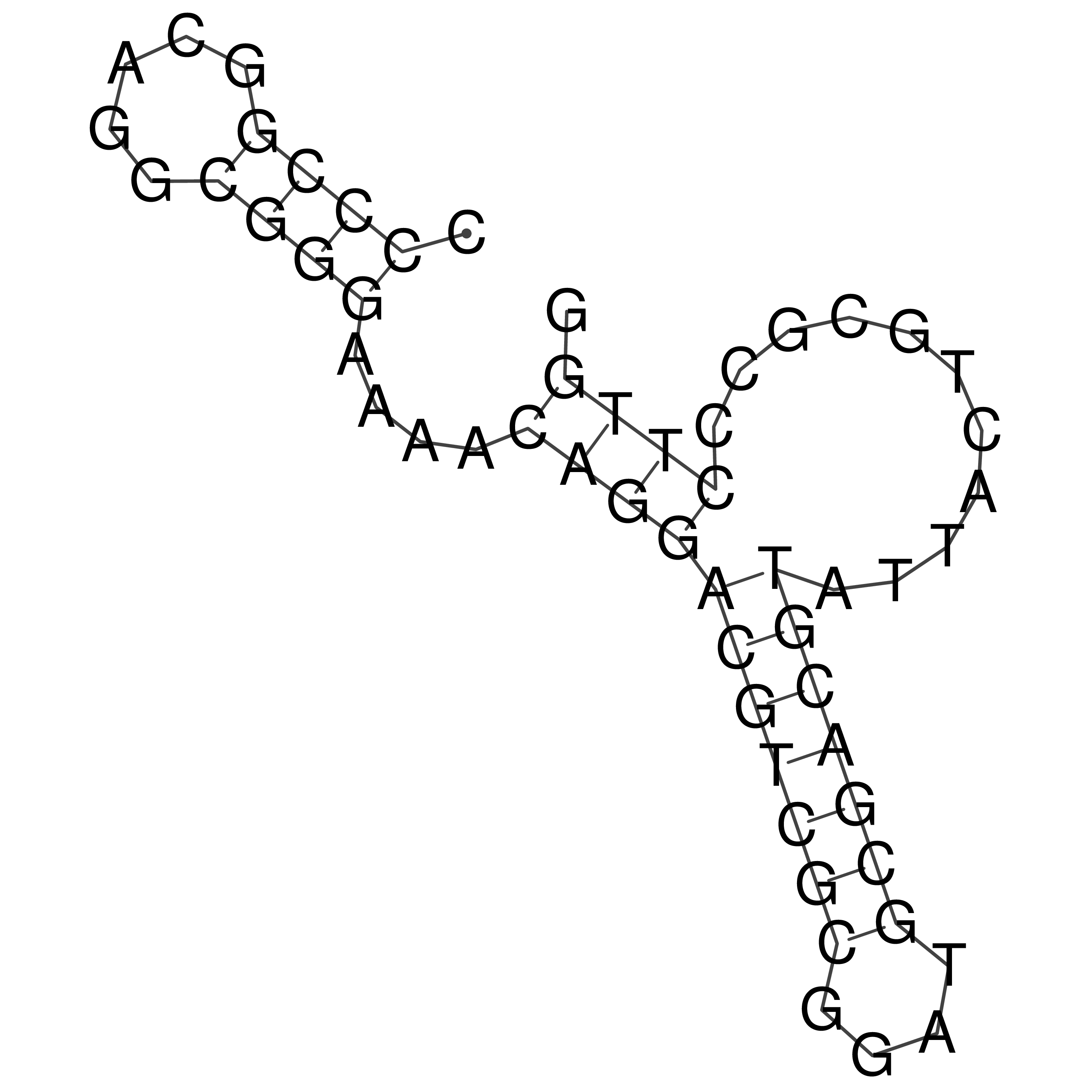
T4CP
| ID | 3615 | GenBank | WP_176540777 |
| Name | t4cp2_HU675_RS50700_pBs162S2c |
UniProt ID | _ |
| Length | 817 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 817 a.a. Molecular weight: 91346.76 Da Isoelectric Point: 6.3242
>WP_176540777.1 conjugal transfer protein TrbE [Bradyrhizobium septentrionale]
MVALHSFRHSGPSFADLVPYAGLVDDGIVLLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILARLG
SGWMIQVEALRIPTTDYPARKDGHFPDAVTREIDEERRAHFERESGHYESRHALILSWRPPERRKSGLAK
YVYSDKDSRSASFADTALGNFRASVREIEQYLSNLLSIQRMRTRTVEERGGSRSARYDELFQFVRFCITG
ENHPVRLPEVPMYLDWLVTAEYQHGLTPTVDGRYVGVVAIDGLPAESWPGILNVLDQMPLTYRWSSRFMF
LDEIEARERLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVAETQDAIAEAASQLVAYGYYTPVIVL
QDHDADRLREKCEGIRRVVQAEGFGARIETLNATEAFLGSLPGNWYANIREPLIHTRNLADLLPLNSVWS
GSPVAPCPFYPESSPPLMQVASGSTPFRLNLHTDDVGHSLIFGPTGSGKSTLLALIAAQFRRYRDAQIFA
FDKGRSLMPLTLAAGGDHYEIGGGAGEGLQLAFCPLAELATDADRAWASEWIETLVVLQGVSVTPDHRNA
ISRQVTLMAAAPGRSLADFVSGVQMREIKDALHHYTVDGPMGHLLDAERDGLTLGGFQTFEIEELMNMGE
RSLVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERLGFNERQVEIVSNAIPKREYYVTSPAGRRLFEMALGPV
ALAFVGASGRDDLKRITELKTAHGRTWPANWLQTRGIAHADTLLAHD
MVALHSFRHSGPSFADLVPYAGLVDDGIVLLKDGSLMAGWYFAGPDSESSTDAERNDVSRQINAILARLG
SGWMIQVEALRIPTTDYPARKDGHFPDAVTREIDEERRAHFERESGHYESRHALILSWRPPERRKSGLAK
YVYSDKDSRSASFADTALGNFRASVREIEQYLSNLLSIQRMRTRTVEERGGSRSARYDELFQFVRFCITG
ENHPVRLPEVPMYLDWLVTAEYQHGLTPTVDGRYVGVVAIDGLPAESWPGILNVLDQMPLTYRWSSRFMF
LDEIEARERLERTRKKWQQKVRPFFDQLFQTQSRSIDQDAMAMVAETQDAIAEAASQLVAYGYYTPVIVL
QDHDADRLREKCEGIRRVVQAEGFGARIETLNATEAFLGSLPGNWYANIREPLIHTRNLADLLPLNSVWS
GSPVAPCPFYPESSPPLMQVASGSTPFRLNLHTDDVGHSLIFGPTGSGKSTLLALIAAQFRRYRDAQIFA
FDKGRSLMPLTLAAGGDHYEIGGGAGEGLQLAFCPLAELATDADRAWASEWIETLVVLQGVSVTPDHRNA
ISRQVTLMAAAPGRSLADFVSGVQMREIKDALHHYTVDGPMGHLLDAERDGLTLGGFQTFEIEELMNMGE
RSLVPVLTYLFRRIEKRLTGAPSLILLDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDAERS
GIIDVLKESCPTKICLPNGAAREPGTREFYERLGFNERQVEIVSNAIPKREYYVTSPAGRRLFEMALGPV
ALAFVGASGRDDLKRITELKTAHGRTWPANWLQTRGIAHADTLLAHD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 103083..112454
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HU675_RS50645 (HU675_0050645) | 98556..99572 | - | 1017 | WP_225024896 | integrase core domain-containing protein | - |
| HU675_RS50650 (HU675_0050650) | 100066..101400 | + | 1335 | WP_176540782 | IS1182 family transposase | - |
| HU675_RS50655 (HU675_0050655) | 101453..101611 | + | 159 | Protein_84 | IS256 family transposase | - |
| HU675_RS50660 (HU675_0050660) | 101590..101823 | - | 234 | Protein_85 | hypothetical protein | - |
| HU675_RS50665 (HU675_0050665) | 101901..102611 | - | 711 | WP_166107311 | autoinducer binding domain-containing protein | - |
| HU675_RS50670 (HU675_0050670) | 103083..104378 | - | 1296 | WP_166107234 | IncP-type conjugal transfer protein TrbI | virB10 |
| HU675_RS50675 (HU675_0050675) | 104392..104856 | - | 465 | WP_166107236 | conjugal transfer protein TrbH | - |
| HU675_RS50680 (HU675_0050680) | 104861..105685 | - | 825 | WP_166107238 | P-type conjugative transfer protein TrbG | virB9 |
| HU675_RS50685 (HU675_0050685) | 105703..106365 | - | 663 | WP_176540780 | conjugal transfer protein TrbF | virB8 |
| HU675_RS50690 (HU675_0050690) | 106381..107568 | - | 1188 | WP_176540779 | P-type conjugative transfer protein TrbL | virB6 |
| HU675_RS50695 (HU675_0050695) | 107562..108380 | - | 819 | WP_176540778 | P-type conjugative transfer protein TrbJ | virB5 |
| HU675_RS50700 (HU675_0050700) | 108352..110805 | - | 2454 | WP_176540777 | conjugal transfer protein TrbE | virb4 |
| HU675_RS50705 (HU675_0050705) | 110815..111114 | - | 300 | WP_176540776 | conjugal transfer protein TrbD | virB3 |
| HU675_RS50710 (HU675_0050710) | 111107..111496 | - | 390 | WP_166218034 | TrbC/VirB2 family protein | virB2 |
| HU675_RS50715 (HU675_0050715) | 111486..112454 | - | 969 | WP_176540775 | P-type conjugative transfer ATPase TrbB | virB11 |
| HU675_RS50720 (HU675_0050720) | 112451..113086 | - | 636 | WP_176540774 | acyl-homoserine-lactone synthase | - |
| HU675_RS50725 (HU675_0050725) | 113447..114682 | + | 1236 | WP_175618600 | plasmid partitioning protein RepA | - |
| HU675_RS50730 (HU675_0050730) | 114858..115871 | + | 1014 | WP_176540773 | plasmid partitioning protein RepB | - |
| HU675_RS50735 (HU675_0050735) | 116087..117406 | + | 1320 | WP_176540772 | plasmid replication protein RepC | - |
Host bacterium
| ID | 5779 | GenBank | NZ_CP088291 |
| Plasmid name | pBs162S2c | Incompatibility group | - |
| Plasmid size | 177103 bp | Coordinate of oriT [Strand] | 9113..9168 [-] |
| Host baterium | Bradyrhizobium septentrionale strain 162S2 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |