Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105307 |
| Name | oriT_RHBSTW-00409|unnamed |
| Organism | Klebsiella michiganensis strain RHBSTW-00409 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055327 (172330..172378 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_RHBSTW-00409|unnamed
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file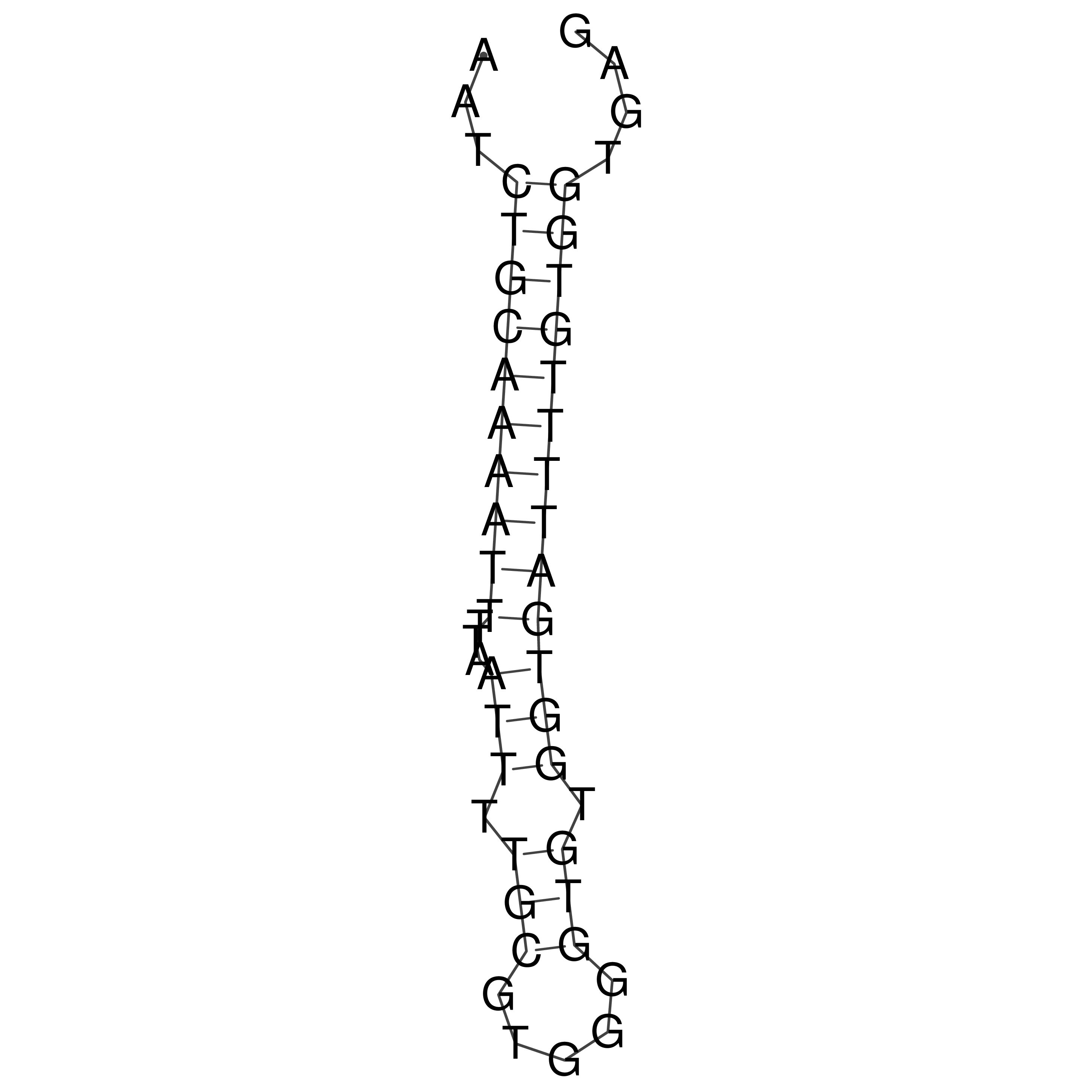
Relaxase
| ID | 3627 | GenBank | WP_048235021 |
| Name | traI_HV187_RS30800_RHBSTW-00409|unnamed |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190795.84 Da Isoelectric Point: 6.1046
>WP_048235021.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGSWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRLALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQFVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRLVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPENRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKDAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQR
SAADVAIMKEIVRQMPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGSWAPGSSVVEFTPKQEKAIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRLALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQFVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3596 | GenBank | WP_048235023 |
| Name | traD_HV187_RS30805_RHBSTW-00409|unnamed |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85941.02 Da Isoelectric Point: 5.1149
>WP_048235023.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRSTEPSALRLTTVPLIKPKAAAAAAAAATVSSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDNVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRSTEPSALRLTTVPLIKPKAAAAAAAAATVSSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDNVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 144472..172936
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV187_RS30805 (HV187_30815) | 144472..146784 | - | 2313 | WP_048235023 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HV187_RS32220 | 146840..146953 | - | 114 | Protein_142 | hypothetical protein | - |
| HV187_RS30810 (HV187_30820) | 147144..147875 | - | 732 | WP_032415736 | conjugal transfer complement resistance protein TraT | - |
| HV187_RS30815 (HV187_30825) | 148127..148729 | - | 603 | WP_023280878 | hypothetical protein | - |
| HV187_RS30820 (HV187_30830) | 148753..151575 | - | 2823 | WP_048235025 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV187_RS30825 (HV187_30835) | 151575..152945 | - | 1371 | WP_048235027 | conjugal transfer pilus assembly protein TraH | traH |
| HV187_RS30830 (HV187_30840) | 152932..153354 | - | 423 | WP_049010971 | hypothetical protein | - |
| HV187_RS30835 (HV187_30845) | 153347..153919 | - | 573 | WP_048235029 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV187_RS30840 (HV187_30850) | 153891..154130 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV187_RS30845 (HV187_30855) | 154141..154893 | - | 753 | WP_048235030 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV187_RS30850 (HV187_30860) | 154914..155240 | - | 327 | WP_048235032 | hypothetical protein | - |
| HV187_RS30855 (HV187_30865) | 155253..155501 | - | 249 | WP_004152675 | hypothetical protein | - |
| HV187_RS30860 (HV187_30870) | 155479..155733 | - | 255 | WP_020804203 | conjugal transfer protein TrbE | - |
| HV187_RS30865 (HV187_30875) | 155765..157720 | - | 1956 | WP_177340687 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV187_RS30870 (HV187_30880) | 157779..158417 | - | 639 | WP_048235035 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV187_RS30875 (HV187_30885) | 158430..159419 | - | 990 | WP_013214027 | conjugal transfer pilus assembly protein TraU | traU |
| HV187_RS30880 (HV187_30890) | 159416..159817 | - | 402 | WP_004194979 | hypothetical protein | - |
| HV187_RS30885 (HV187_30895) | 159852..160487 | - | 636 | WP_048235038 | type-F conjugative transfer system protein TraW | traW |
| HV187_RS30890 (HV187_30900) | 160487..160876 | - | 390 | WP_048235039 | type-F conjugative transfer system protein TrbI | - |
| HV187_RS30895 (HV187_30905) | 160876..163515 | - | 2640 | WP_004197813 | type IV secretion system protein TraC | virb4 |
| HV187_RS30900 (HV187_30910) | 163587..163985 | - | 399 | WP_074194504 | hypothetical protein | - |
| HV187_RS30905 (HV187_30915) | 164363..164767 | - | 405 | WP_004197817 | hypothetical protein | - |
| HV187_RS30910 (HV187_30920) | 164834..165145 | - | 312 | WP_025712701 | hypothetical protein | - |
| HV187_RS30915 (HV187_30925) | 165146..165364 | - | 219 | WP_004195468 | hypothetical protein | - |
| HV187_RS32225 | 165388..165636 | - | 249 | WP_223175793 | hypothetical protein | - |
| HV187_RS32230 | 165707..166105 | - | 399 | WP_020277946 | hypothetical protein | - |
| HV187_RS30925 (HV187_30935) | 166260..166844 | - | 585 | WP_020277945 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV187_RS30930 (HV187_30940) | 166918..168342 | - | 1425 | WP_015065626 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV187_RS30935 (HV187_30945) | 168342..169082 | - | 741 | WP_048235042 | type-F conjugative transfer system secretin TraK | traK |
| HV187_RS30940 (HV187_30950) | 169069..169635 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| HV187_RS30945 (HV187_30955) | 169655..169960 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HV187_RS30950 (HV187_30960) | 169974..170342 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| HV187_RS30955 (HV187_30965) | 170410..170610 | - | 201 | WP_046664192 | TraY domain-containing protein | - |
| HV187_RS30960 (HV187_30970) | 170696..171397 | - | 702 | WP_025712700 | hypothetical protein | - |
| HV187_RS30965 (HV187_30975) | 171627..172019 | - | 393 | WP_004194114 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV187_RS30970 (HV187_30980) | 172451..172936 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HV187_RS30975 (HV187_30985) | 172969..173298 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| HV187_RS30980 (HV187_30990) | 173331..174152 | - | 822 | WP_023316395 | DUF932 domain-containing protein | - |
| HV187_RS30985 (HV187_30995) | 175074..176363 | - | 1290 | WP_004574636 | ISL3-like element ISPpu12 family transposase | - |
| HV187_RS30990 (HV187_31000) | 176385..176897 | - | 513 | WP_004863699 | signal peptidase II | - |
| HV187_RS30995 (HV187_31005) | 176901..177797 | - | 897 | WP_004574643 | cation transporter | - |
Host bacterium
| ID | 5745 | GenBank | NZ_CP055327 |
| Plasmid name | RHBSTW-00409|unnamed | Incompatibility group | IncFIB |
| Plasmid size | 215915 bp | Coordinate of oriT [Strand] | 172330..172378 [+] |
| Host baterium | Klebsiella michiganensis strain RHBSTW-00409 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsR, arsD, arsA, arsB, arsC, arsH, pcoE, pcoS, pcoR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE, fecD, fecE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |