Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105301 |
| Name | oriT_pRHB33-C07_2 |
| Organism | Escherichia fergusonii strain RHB33-C07 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP057211 (117147..117232 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
AATTACATGATTTAAAACGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file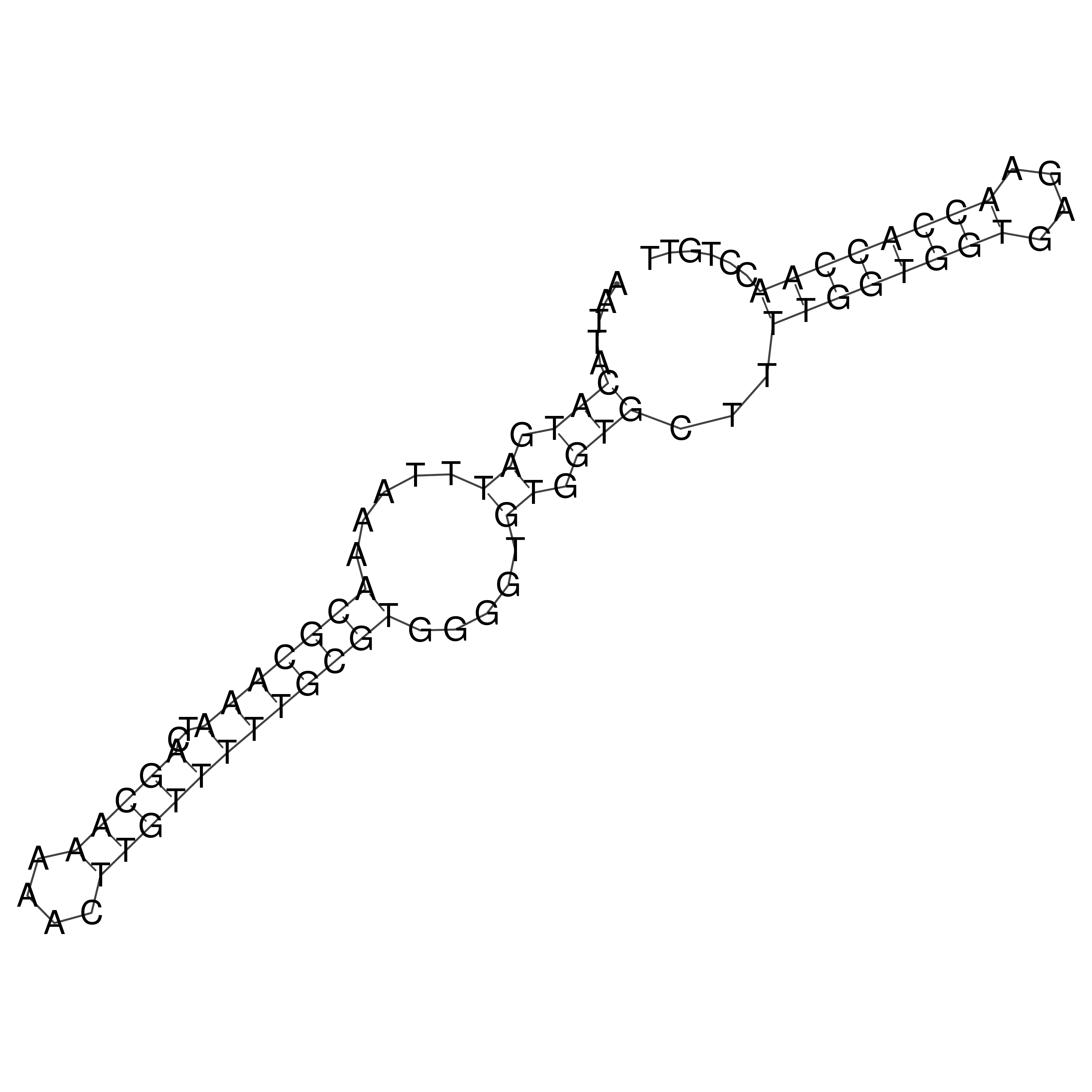
Auxiliary protein
| ID | 1626 | GenBank | WP_040100321 |
| Name | WP_040100321_pRHB33-C07_2 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14508.47 Da Isoelectric Point: 4.7033
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISLSGTASMLLELGLRVYDAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1627 | GenBank | WP_072039828 |
| Name | WP_072039828_pRHB33-C07_2 |
UniProt ID | _ |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8515.70 Da Isoelectric Point: 10.3461
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLRRFPDFYNIDSIKEGAGETDS
TIKDI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3589 | GenBank | WP_105466876 |
| Name | traC_HVZ09_RS23660_pRHB33-C07_2 |
UniProt ID | _ |
| Length | 881 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 881 a.a. Molecular weight: 99900.68 Da Isoelectric Point: 6.1932
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANESIVEALDHMLRTKLPRGVPFCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMNAA
ATQFPLPEGMNLPLTLRHYRVFFSYCSPSKKKSRADILEMENLVKIIRASLQGASITTQAVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKKNRARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQY
TAGGTYGRYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRNLKKLNVIDEGW
RLLDFKNHKVGEFIEKGYRTARRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLYPDQFLPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRKEG
LSIHEAVWQLAWKKSGPEMASLEAWLKEHEMSGERMTGNWS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3590 | GenBank | WP_181667511 |
| Name | traD_HVZ09_RS23750_pRHB33-C07_2 |
UniProt ID | _ |
| Length | 720 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 720 a.a. Molecular weight: 81854.49 Da Isoelectric Point: 5.1126
MSFSAKDMTQGGQIASMRIRMFSQIANIILYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIRSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEITGGRQLTDNPKEVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQTRPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPVS
PAINDKKSDAGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAVYEAWQREQNPDIQQKMQRRE
EVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 116579..148435
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HVZ09_RS23540 (HVZ09_23490) | 112570..113004 | + | 435 | WP_032235533 | conjugation system SOS inhibitor PsiB | - |
| HVZ09_RS23545 (HVZ09_23495) | 113001..113763 | + | 763 | Protein_114 | plasmid SOS inhibition protein A | - |
| HVZ09_RS24715 | 113936..114088 | + | 153 | Protein_115 | DUF5431 family protein | - |
| HVZ09_RS23550 (HVZ09_23500) | 114033..114155 | + | 123 | WP_223170659 | type I toxin-antitoxin system Hok family toxin | - |
| HVZ09_RS24520 (HVZ09_23505) | 114456..114753 | - | 298 | Protein_117 | hypothetical protein | - |
| HVZ09_RS23555 (HVZ09_23510) | 114824..115030 | + | 207 | WP_181667506 | single-stranded DNA-binding protein | - |
| HVZ09_RS23560 (HVZ09_23515) | 115055..115342 | + | 288 | WP_021569031 | hypothetical protein | - |
| HVZ09_RS23565 (HVZ09_23520) | 115461..116282 | + | 822 | WP_042973148 | DUF932 domain-containing protein | - |
| HVZ09_RS23570 (HVZ09_23525) | 116579..117181 | - | 603 | WP_072156291 | transglycosylase SLT domain-containing protein | virB1 |
| HVZ09_RS23575 (HVZ09_23530) | 117505..117888 | + | 384 | WP_040100321 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HVZ09_RS23580 (HVZ09_23535) | 118083..118754 | + | 672 | WP_040100314 | conjugal transfer transcriptional regulator TraJ | - |
| HVZ09_RS23585 (HVZ09_23540) | 118892..119119 | + | 228 | WP_072039828 | conjugal transfer relaxosome protein TraY | - |
| HVZ09_RS23590 (HVZ09_23545) | 119153..119518 | + | 366 | WP_040100313 | type IV conjugative transfer system pilin TraA | - |
| HVZ09_RS23595 (HVZ09_23550) | 119533..119844 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HVZ09_RS23600 (HVZ09_23555) | 119866..120432 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HVZ09_RS23605 (HVZ09_23560) | 120419..121147 | + | 729 | WP_202117066 | type-F conjugative transfer system secretin TraK | traK |
| HVZ09_RS23610 (HVZ09_23565) | 121147..122574 | + | 1428 | WP_000146635 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HVZ09_RS23615 (HVZ09_23570) | 122564..123154 | + | 591 | WP_000002784 | conjugal transfer pilus-stabilizing protein TraP | - |
| HVZ09_RS23620 (HVZ09_23575) | 123141..123461 | + | 321 | WP_249418243 | conjugal transfer protein TrbD | - |
| HVZ09_RS23625 (HVZ09_23580) | 123458..123973 | + | 516 | WP_105466871 | type IV conjugative transfer system lipoprotein TraV | traV |
| HVZ09_RS23630 (HVZ09_23585) | 124107..124328 | + | 222 | WP_105466872 | conjugal transfer protein TraR | - |
| HVZ09_RS23635 (HVZ09_23590) | 124473..125219 | - | 747 | WP_001016257 | IS21-like element ISEc12 family helper ATPase IstB | - |
| HVZ09_RS23640 (HVZ09_23595) | 125234..126775 | - | 1542 | WP_001298859 | IS21-like element ISEc12 family transposase | - |
| HVZ09_RS23645 (HVZ09_23600) | 126956..127381 | + | 426 | WP_181667507 | hypothetical protein | - |
| HVZ09_RS24720 | 127461..127678 | + | 218 | Protein_137 | hypothetical protein | - |
| HVZ09_RS23650 (HVZ09_23605) | 127675..128079 | + | 405 | WP_105466874 | hypothetical protein | - |
| HVZ09_RS23655 (HVZ09_23610) | 128076..128423 | + | 348 | WP_105466875 | hypothetical protein | - |
| HVZ09_RS23660 (HVZ09_23615) | 128549..131194 | + | 2646 | WP_105466876 | type IV secretion system protein TraC | virb4 |
| HVZ09_RS23665 (HVZ09_23620) | 131576..132682 | - | 1107 | WP_181667508 | hypothetical protein | - |
| HVZ09_RS23670 (HVZ09_23625) | 132778..133146 | + | 369 | WP_249418244 | type-F conjugative transfer system protein TrbI | - |
| HVZ09_RS23675 (HVZ09_23630) | 133143..133775 | + | 633 | WP_181667509 | type-F conjugative transfer system protein TraW | traW |
| HVZ09_RS23680 (HVZ09_23635) | 133772..134764 | + | 993 | WP_040100302 | conjugal transfer pilus assembly protein TraU | traU |
| HVZ09_RS23685 (HVZ09_23640) | 134773..135411 | + | 639 | WP_000777692 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HVZ09_RS23690 (HVZ09_23645) | 135408..137216 | + | 1809 | WP_040100300 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HVZ09_RS23695 (HVZ09_23650) | 137243..137503 | + | 261 | WP_000864348 | conjugal transfer protein TrbE | - |
| HVZ09_RS23700 (HVZ09_23655) | 137496..138239 | + | 744 | WP_181667534 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HVZ09_RS23705 (HVZ09_23660) | 138400..138636 | + | 237 | WP_000270010 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HVZ09_RS23710 (HVZ09_23665) | 138623..139168 | + | 546 | WP_000059821 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HVZ09_RS23715 (HVZ09_23670) | 139158..139438 | + | 281 | Protein_151 | P-type conjugative transfer protein TrbJ | - |
| HVZ09_RS23720 (HVZ09_23675) | 139425..139805 | + | 381 | WP_249540839 | F-type conjugal transfer protein TrbF | - |
| HVZ09_RS23725 (HVZ09_23680) | 139805..141178 | + | 1374 | WP_181667510 | conjugal transfer pilus assembly protein TraH | traH |
| HVZ09_RS23730 (HVZ09_23685) | 141175..143994 | + | 2820 | WP_151674034 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HVZ09_RS23735 (HVZ09_23690) | 144019..144516 | + | 498 | WP_137546094 | surface exclusion protein | - |
| HVZ09_RS23740 (HVZ09_23695) | 144548..145279 | + | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| HVZ09_RS23745 (HVZ09_23700) | 145482..146222 | + | 741 | WP_151674032 | hypothetical protein | - |
| HVZ09_RS23750 (HVZ09_23705) | 146273..148435 | + | 2163 | WP_181667511 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HVZ09_RS23755 (HVZ09_23710) | 148444..148842 | - | 399 | WP_000911313 | type II toxin-antitoxin system VapC family toxin | - |
| HVZ09_RS23760 (HVZ09_23715) | 148842..149069 | - | 228 | WP_000450520 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 5739 | GenBank | NZ_CP057211 |
| Plasmid name | pRHB33-C07_2 | Incompatibility group | IncFIB |
| Plasmid size | 160081 bp | Coordinate of oriT [Strand] | 117147..117232 [-] |
| Host baterium | Escherichia fergusonii strain RHB33-C07 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | pefA, pefC, pefD |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |