Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105288 |
| Name | oriT_pRHB35-C21_2 |
| Organism | Citrobacter sp. RHB35-C21 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP057148 (170887..171171 [+], 285 nt) |
| oriT length | 285 nt |
| IRs (inverted repeats) | 184..189, 191..196 (AAAAGT..ACTTTT) |
| Location of nic site | 109..110 |
| Conserved sequence flanking the nic site |
TTTGGTTAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 285 nt
>oriT_pRHB35-C21_2
GGTTATTGCTACTTAATGCCGATAACGACTCAGGCTTTGAGGTTTTTTTATACGGTTCACATTTCGTTAGCAAGGTCAGGGTTTTTTGATAAAATTCTGGTTAGTTTGGTTAAAAAGTGTTACAAGTATGGGTAATGGCTGAAAGGTTAGTTTTAAGGTTCAAAGCGGCAGTATTAAAATTCCAAAAGTTACTTTTCATCCTTCAGAATCCAGACCTTAATTTCATGTAGAAGATTCGTACAATTGTATTGGCGCAAGGACAATCCGCACATGTCAGAATCAGAT
GGTTATTGCTACTTAATGCCGATAACGACTCAGGCTTTGAGGTTTTTTTATACGGTTCACATTTCGTTAGCAAGGTCAGGGTTTTTTGATAAAATTCTGGTTAGTTTGGTTAAAAAGTGTTACAAGTATGGGTAATGGCTGAAAGGTTAGTTTTAAGGTTCAAAGCGGCAGTATTAAAATTCCAAAAGTTACTTTTCATCCTTCAGAATCCAGACCTTAATTTCATGTAGAAGATTCGTACAATTGTATTGGCGCAAGGACAATCCGCACATGTCAGAATCAGAT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file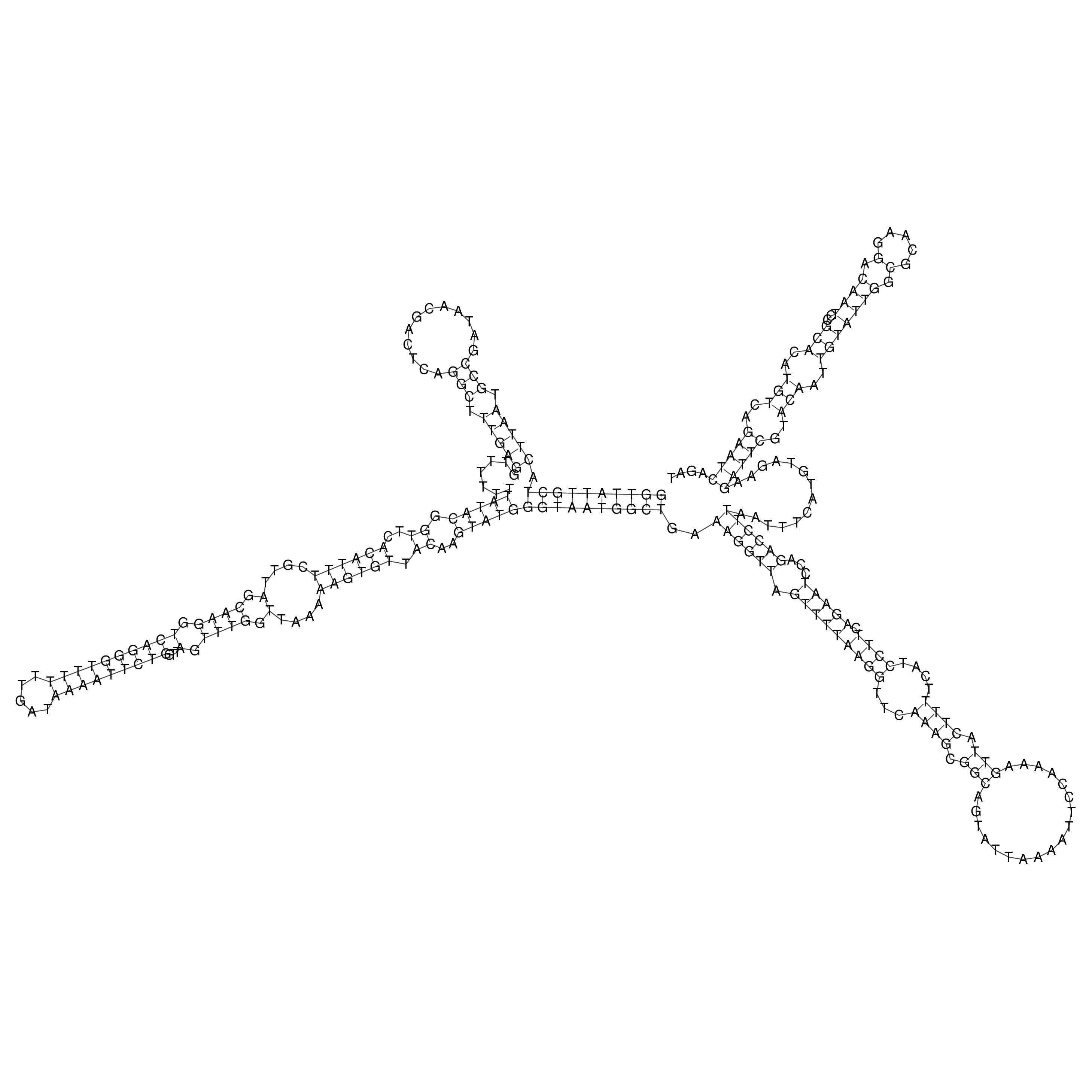
Host bacterium
| ID | 5726 | GenBank | NZ_CP057148 |
| Plasmid name | pRHB35-C21_2 | Incompatibility group | - |
| Plasmid size | 252642 bp | Coordinate of oriT [Strand] | 170887..171171 [+] |
| Host baterium | Citrobacter sp. RHB35-C21 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsR, arsB, arsC, fecE, fecD, yqjH, arsA, arsD, pcoE, pcoS, pcoR, pcoD, pcoC, pcoB, pcoA, silP, silA, silB, silF, silC, silR, silS, silE, merR, merT, merP, merA, merD, merE, nirD, ncrC, ncrB, ncrA |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |