Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105275 |
| Name | oriT_pRHB23-C01_5 |
| Organism | Escherichia fergusonii strain RHB23-C01 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP057569 (16909..17011 [-], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 65..70, 73..78 (CACCGT..ACGGTG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_pRHB23-C01_5
AGCCTCGCAGAGCAGGACTCCCGTTGAGCAGCCCCGCTGCGAATAAGGGAAGGTAAAGTAGAAGCACCGTTTACGGTGGGCCTACTTTACATGTCCTGCCCGG
AGCCTCGCAGAGCAGGACTCCCGTTGAGCAGCCCCGCTGCGAATAAGGGAAGGTAAAGTAGAAGCACCGTTTACGGTGGGCCTACTTTACATGTCCTGCCCGG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file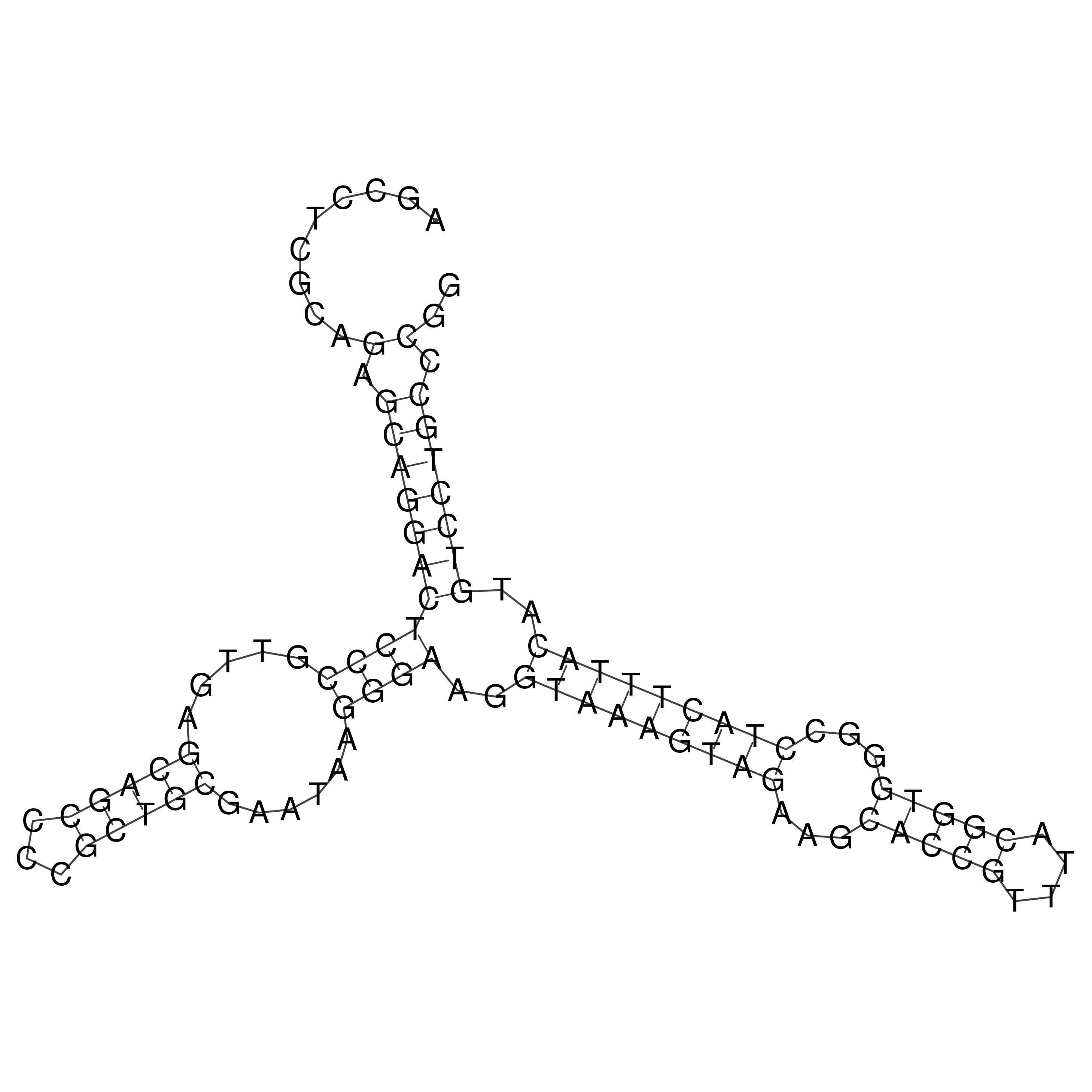
Relaxase
| ID | 3611 | GenBank | WP_023352245 |
| Name | Relaxase_HVX91_RS23410_pRHB23-C01_5 |
UniProt ID | _ |
| Length | 877 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 877 a.a. Molecular weight: 97961.42 Da Isoelectric Point: 10.5942
>WP_023352245.1 MULTISPECIES: TraI/MobA(P) family conjugative relaxase [Enterobacteriaceae]
MISRHIPMRNVKKSSFFRLAEYLQDSQNKQERVGAVKTTNCHQENILDAARYEIEPTQRLNTRAKGDKTY
HLVISFRPGENPPQEVLDVIEERMCTALGYGEHQRVSAVHYDTDNVHIQIAINKVHPKTHNIHTPYNDYK
IRTAVCQELEKEFGLEKDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHKAMRQ
NGLELRQKGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRLSPEAIKQRFGSAVHV
KRPGERPPPIRENRQVHLGEIPVMSINNGKRYEGKPLGSQGASTSVLYGRYQSEQQRLTAARNEALRLAK
VKRDRLIENAKRTGRLKRAAIKLLKGPGVNKRHLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWA
DWLQFKAAGGDDEALKILRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDG
GKRLDVSRDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFTDPELEKLRQILAKK
AAREREYIHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTEA
GRERSGNGNATGANEPGVTGRSPSADAGNGYGTSNSGRRTGSGDGVTTAGSQPGRSDDTRTGRSRAAESN
GERGHRKPNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLPGNVPDQLEHKTAERTDQLRRDMAGA
GGITTPAEVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIV
VMPVSDYTARRLSNLKVGESVTLTPSGSVRTTKGRSR
MISRHIPMRNVKKSSFFRLAEYLQDSQNKQERVGAVKTTNCHQENILDAARYEIEPTQRLNTRAKGDKTY
HLVISFRPGENPPQEVLDVIEERMCTALGYGEHQRVSAVHYDTDNVHIQIAINKVHPKTHNIHTPYNDYK
IRTAVCQELEKEFGLEKDNHRASKTRSENNAADMERHSGRESLLGWIKRECADQLKSAANWQELHKAMRQ
NGLELRQKGNGFVITGPDGLAVKASSVDRSLSRAALEKKLGSFESSIPDTKPSRLSPEAIKQRFGSAVHV
KRPGERPPPIRENRQVHLGEIPVMSINNGKRYEGKPLGSQGASTSVLYGRYQSEQQRLTAARNEALRLAK
VKRDRLIENAKRTGRLKRAAIKLLKGPGVNKRHLYGLASKALGEELERANAIYKKDRQAAYDIHKRRTWA
DWLQFKAAGGDDEALKILRQRNAQQARKEANILHGDKVKKDGRIPGLKPDSVTKEGTIIYRVGESAIRDG
GKRLDVSRDAGDDGLHAALLMAKHRYGDKLTVNGSFEFKKRIVAVAAARELDVSFTDPELEKLRQILAKK
AAREREYIHIPFEQRGKARQAGAMWDTEKKSWFVGPYATRDRIENFIRMNEEKDNEQRRQRYAGTERTEA
GRERSGNGNATGANEPGVTGRSPSADAGNGYGTSNSGRRTGSGDGVTTAGSQPGRSDDTRTGRSRAAESN
GERGHRKPNVARPGRQPPPTAKNRLRAMSELGMVRIAGGSEVLLPGNVPDQLEHKTAERTDQLRRDMAGA
GGITTPAEVKKPLSSAEKYINEREQKRLKGFDIPKHRGYNSTDKGLFEYAGQRKSGLETLALLKRDDEIV
VMPVSDYTARRLSNLKVGESVTLTPSGSVRTTKGRSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3574 | GenBank | WP_096217383 |
| Name | t4cp2_HVX91_RS23420_pRHB23-C01_5 |
UniProt ID | _ |
| Length | 647 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 647 a.a. Molecular weight: 71689.89 Da Isoelectric Point: 8.3842
>WP_096217383.1 MULTISPECIES: type IV secretory system conjugative DNA transfer family protein [Escherichia]
MRKPNNAVGPQVRNKKKAAKGKLIPAMAITSLVAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHERLPDAFFAAGSVGAAVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIRDAGLLNDEGVY
VGAWEDKNGQLRYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYAK
NLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVLY
KHKNEGTPATLPYVDSIMADPERGAGELWMEMTQYSHINGENHPVVGAAGRDMMDRPEEEAGSVLSTAKS
YLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERVD
NNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLGMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQDI
NQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTIQ
EVQRPLLTVDECLRMPGPKKNAEGLITERGDMVIYVAGFPAIYGKQPLFFQDEIFSLRASVPAPKVTDRI
RAVAAANDDASNEAIAI
MRKPNNAVGPQVRNKKKAAKGKLIPAMAITSLVAGIQAATQYFAYSFNYQELLGPNFNHIYAPWSYFKWY
SAWHERLPDAFFAAGSVGAAVAAGGLVLTALTSMMKANSSKANEYLHGSARWADEQDIRDAGLLNDEGVY
VGAWEDKNGQLRYLRHNGPEHVLTYAPTRSGKGVGLVVPTLLSWKHSTVVADLKGELWAMTAGWRKEYAK
NLCLKFEPAAANGSVAWNPLDEIRVGTENEVGDVQNLATLIVDPDGKGLNDHWQKTSQALLVGVILHVLY
KHKNEGTPATLPYVDSIMADPERGAGELWMEMTQYSHINGENHPVVGAAGRDMMDRPEEEAGSVLSTAKS
YLALFRDPVVARNVSESHFKIKDLMNHDDPVSLYIVTQPNDKARLRPLVRIMLNMIVRLLADKMEFERVD
NNLTLWQRFKQAFGFSVQGTKRVQTKKTYKHRLLGMIDEFPSLGKLEIIQESLAFLAGYGIKFYLICQDI
NQLRSRETGYGPDETITSNCHVQNAYPPNRTETAEHLSKLTGQTTVIKEQITTSGKRASAILGGVSKTIQ
EVQRPLLTVDECLRMPGPKKNAEGLITERGDMVIYVAGFPAIYGKQPLFFQDEIFSLRASVPAPKVTDRI
RAVAAANDDASNEAIAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 5713 | GenBank | NZ_CP057569 |
| Plasmid name | pRHB23-C01_5 | Incompatibility group | IncP1 |
| Plasmid size | 41486 bp | Coordinate of oriT [Strand] | 16909..17011 [-] |
| Host baterium | Escherichia fergusonii strain RHB23-C01 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |