Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105263 |
| Name | oriT_pRHB39-C24_3 |
| Organism | Klebsiella pneumoniae strain RHB39-C24 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP057036 (47571..47619 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pRHB39-C24_3
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file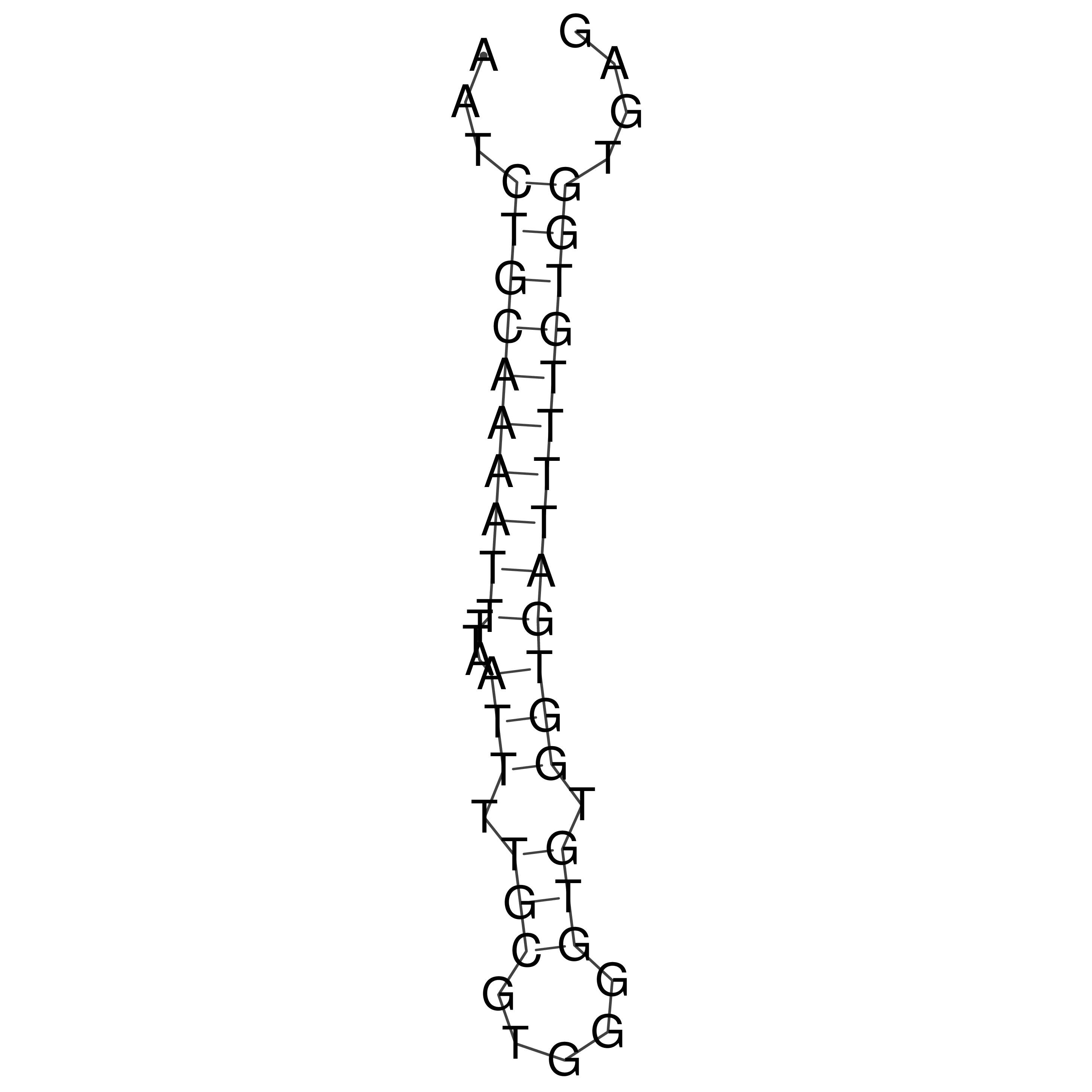
Relaxase
| ID | 3608 | GenBank | WP_075874369 |
| Name | traI_HVZ90_RS26985_pRHB39-C24_3 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190669.51 Da Isoelectric Point: 6.0127
>WP_075874369.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADALPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATR
MILESTDQFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVSSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRLWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANKHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGQQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAV
DSQWTLFRADALPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKTRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTDLTAGQRAATR
MILESTDQFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVSSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKLTRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRLWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3571 | GenBank | WP_077272500 |
| Name | traD_HVZ90_RS26980_pRHB39-C24_3 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85949.93 Da Isoelectric Point: 5.0156
>WP_077272500.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTTKRPAAEPSVRATEPSVLRGTTVPLIKPKAAAAAAAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTTKRPAAEPSVRATEPSVLRGTTVPLIKPKAAAAAAAASTASSAGAPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 47019..75608
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HVZ90_RS26765 (HVZ90_26770) | 42113..42433 | + | 321 | WP_181578661 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HVZ90_RS26770 (HVZ90_26775) | 42423..42701 | + | 279 | WP_004152721 | helix-turn-helix transcriptional regulator | - |
| HVZ90_RS26775 (HVZ90_26780) | 42702..43115 | + | 414 | WP_004182074 | helix-turn-helix domain-containing protein | - |
| HVZ90_RS26780 (HVZ90_26785) | 43833..44213 | + | 381 | WP_044265040 | hypothetical protein | - |
| HVZ90_RS26785 (HVZ90_26790) | 44279..44626 | + | 348 | WP_044265036 | hypothetical protein | - |
| HVZ90_RS26790 (HVZ90_26795) | 44715..44945 | + | 231 | WP_044265034 | hypothetical protein | - |
| HVZ90_RS26795 (HVZ90_26800) | 44992..45825 | + | 834 | WP_044265032 | N-6 DNA methylase | - |
| HVZ90_RS26800 (HVZ90_26805) | 46029..46259 | - | 231 | WP_044265030 | hypothetical protein | - |
| HVZ90_RS26805 (HVZ90_26810) | 46451..46981 | + | 531 | WP_075874364 | antirestriction protein | - |
| HVZ90_RS26810 (HVZ90_26815) | 47019..47426 | - | 408 | WP_225378279 | transglycosylase SLT domain-containing protein | virB1 |
| HVZ90_RS26815 (HVZ90_26820) | 47930..48322 | + | 393 | WP_004194114 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HVZ90_RS26820 (HVZ90_26825) | 48552..49253 | + | 702 | WP_040120380 | hypothetical protein | - |
| HVZ90_RS26825 (HVZ90_26830) | 49339..49539 | + | 201 | WP_046664192 | TraY domain-containing protein | - |
| HVZ90_RS26830 (HVZ90_26835) | 49607..49975 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| HVZ90_RS26835 (HVZ90_26840) | 49989..50294 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HVZ90_RS26840 (HVZ90_26845) | 50314..50880 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| HVZ90_RS26845 (HVZ90_26850) | 50867..51607 | + | 741 | WP_040120379 | type-F conjugative transfer system secretin TraK | traK |
| HVZ90_RS26850 (HVZ90_26855) | 51607..53031 | + | 1425 | WP_181578653 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HVZ90_RS26855 (HVZ90_26860) | 53104..53688 | + | 585 | WP_040120377 | type IV conjugative transfer system lipoprotein TraV | traV |
| HVZ90_RS26860 (HVZ90_26865) | 54011..54229 | + | 219 | WP_004195468 | hypothetical protein | - |
| HVZ90_RS26865 (HVZ90_26870) | 54230..54541 | + | 312 | WP_029497417 | hypothetical protein | - |
| HVZ90_RS26870 (HVZ90_26875) | 54608..55012 | + | 405 | WP_004197817 | hypothetical protein | - |
| HVZ90_RS27310 | 55055..55207 | + | 153 | WP_224518895 | hypothetical protein | - |
| HVZ90_RS26880 (HVZ90_26885) | 55389..55787 | + | 399 | WP_023179972 | hypothetical protein | - |
| HVZ90_RS26885 (HVZ90_26890) | 55859..58498 | + | 2640 | WP_013214024 | type IV secretion system protein TraC | virb4 |
| HVZ90_RS26890 (HVZ90_26895) | 58498..58887 | + | 390 | WP_004167468 | type-F conjugative transfer system protein TrbI | - |
| HVZ90_RS26895 (HVZ90_26900) | 58887..59522 | + | 636 | WP_029497419 | type-F conjugative transfer system protein TraW | traW |
| HVZ90_RS26900 (HVZ90_26905) | 59557..59958 | + | 402 | WP_004194979 | hypothetical protein | - |
| HVZ90_RS26905 (HVZ90_26910) | 59955..60944 | + | 990 | WP_029497420 | conjugal transfer pilus assembly protein TraU | traU |
| HVZ90_RS26910 (HVZ90_26915) | 60957..61595 | + | 639 | WP_015065635 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HVZ90_RS26915 (HVZ90_26920) | 61654..63609 | + | 1956 | WP_181578654 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HVZ90_RS26920 (HVZ90_26925) | 63641..63895 | + | 255 | WP_044265005 | conjugal transfer protein TrbE | - |
| HVZ90_RS26925 (HVZ90_26930) | 63873..64121 | + | 249 | WP_004152675 | hypothetical protein | - |
| HVZ90_RS26930 (HVZ90_26935) | 64134..64460 | + | 327 | WP_004144402 | hypothetical protein | - |
| HVZ90_RS26935 (HVZ90_26940) | 64481..65233 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HVZ90_RS26940 (HVZ90_26945) | 65244..65483 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HVZ90_RS26945 (HVZ90_26950) | 65455..66012 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HVZ90_RS26950 (HVZ90_26955) | 66058..66501 | + | 444 | WP_014343488 | F-type conjugal transfer protein TrbF | - |
| HVZ90_RS26955 (HVZ90_26960) | 66479..67858 | + | 1380 | WP_072198320 | conjugal transfer pilus assembly protein TraH | traH |
| HVZ90_RS26960 (HVZ90_26965) | 67858..70680 | + | 2823 | WP_075874360 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HVZ90_RS26965 (HVZ90_26970) | 70703..71305 | + | 603 | WP_013214035 | conjugal transfer protein TraS | - |
| HVZ90_RS26970 (HVZ90_26975) | 71556..72287 | + | 732 | WP_016831056 | conjugal transfer complement resistance protein TraT | - |
| HVZ90_RS26975 (HVZ90_26980) | 72480..73169 | + | 690 | WP_014343485 | hypothetical protein | - |
| HVZ90_RS26980 (HVZ90_26985) | 73296..75608 | + | 2313 | WP_077272500 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5701 | GenBank | NZ_CP057036 |
| Plasmid name | pRHB39-C24_3 | Incompatibility group | IncFII |
| Plasmid size | 86316 bp | Coordinate of oriT [Strand] | 47571..47619 [-] |
| Host baterium | Klebsiella pneumoniae strain RHB39-C24 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |