Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105254 |
| Name | oriT_pRHBSTW-00167_2 |
| Organism | Klebsiella michiganensis strain RHBSTW-00167 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP058119 (185268..185316 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pRHBSTW-00167_2
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTTTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTTTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file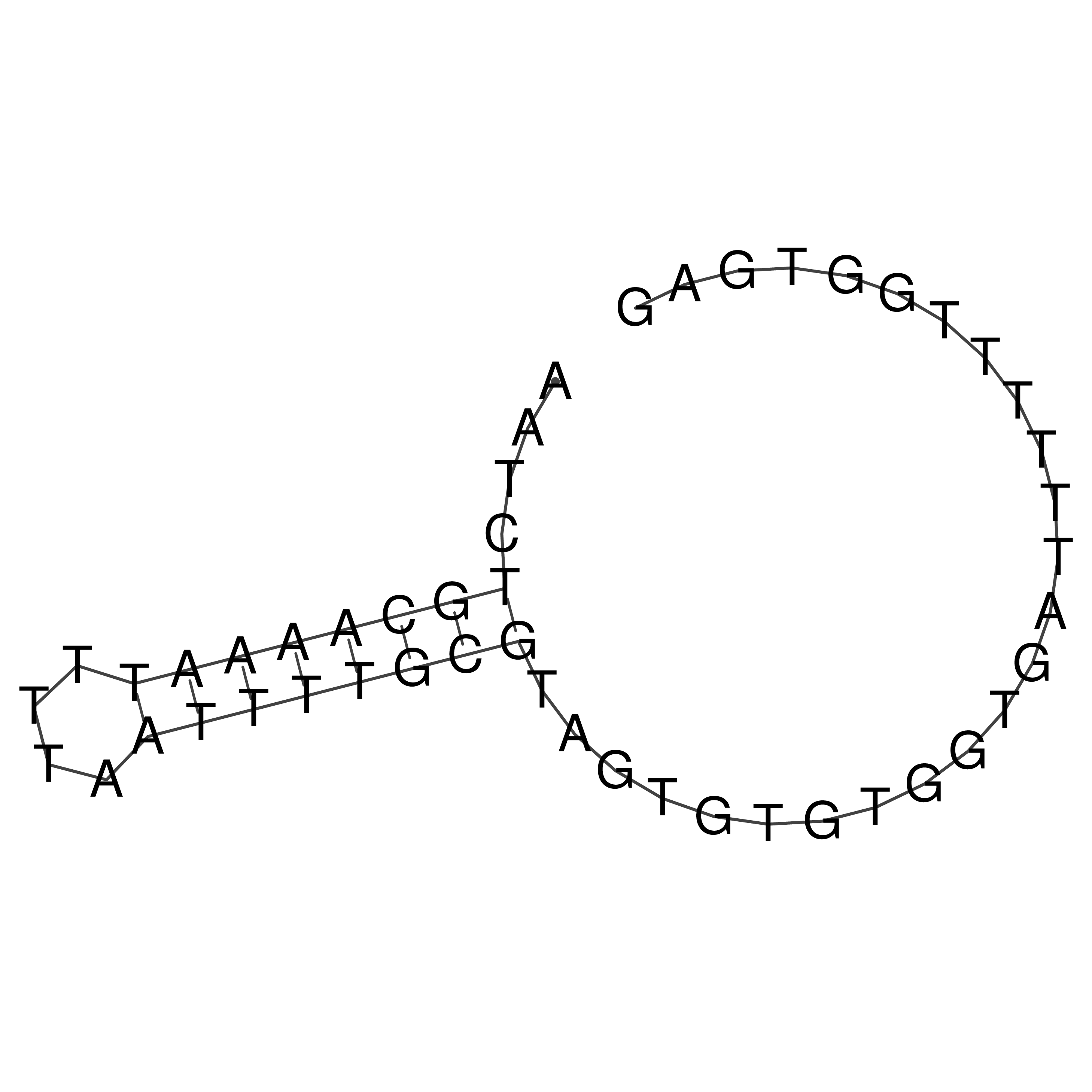
Relaxase
| ID | 3604 | GenBank | WP_181250924 |
| Name | traI_HV105_RS30145_pRHBSTW-00167_2 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190223.95 Da Isoelectric Point: 6.2880
>WP_181250924.1 conjugative transfer relaxase/helicase TraI [Klebsiella michiganensis]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRNEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRAPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRNEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGGRLDLKVSAVD
SQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESITVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
IGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATALETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAHGHGNDLLISRQTWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFTPKQEKAIQK
ALSEGKTLPAGQPATLYEALVKDYTGRAPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDLERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIQHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3567 | GenBank | WP_020316650 |
| Name | traD_HV105_RS30140_pRHBSTW-00167_2 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 85197.99 Da Isoelectric Point: 4.8865
>WP_020316650.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTEQELAQQ
SAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTEQELAQQ
SAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 187695..213220
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV105_RS29960 (HV105_29960) | 184148..184678 | + | 531 | WP_015632491 | antirestriction protein | - |
| HV105_RS29965 (HV105_29965) | 184716..185123 | - | 408 | WP_020805750 | transglycosylase SLT domain-containing protein | - |
| HV105_RS29970 (HV105_29970) | 185627..186019 | + | 393 | WP_032419915 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV105_RS32155 | 186257..186958 | + | 702 | WP_032419917 | hypothetical protein | - |
| HV105_RS29980 (HV105_29980) | 187044..187244 | + | 201 | WP_048289787 | TraY domain-containing protein | - |
| HV105_RS29985 (HV105_29985) | 187313..187681 | + | 369 | WP_020316649 | type IV conjugative transfer system pilin TraA | - |
| HV105_RS29990 (HV105_29990) | 187695..188000 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| HV105_RS29995 (HV105_29995) | 188020..188586 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| HV105_RS30000 (HV105_30000) | 188573..189313 | + | 741 | WP_020316652 | type-F conjugative transfer system secretin TraK | traK |
| HV105_RS30005 (HV105_30005) | 189313..190737 | + | 1425 | WP_020316642 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV105_RS30010 (HV105_30010) | 190730..191326 | + | 597 | WP_023340943 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV105_RS30015 (HV105_30015) | 191349..191918 | + | 570 | WP_032419921 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV105_RS30020 (HV105_30020) | 192050..192460 | + | 411 | WP_032419923 | hypothetical protein | - |
| HV105_RS30025 (HV105_30025) | 192465..192755 | + | 291 | WP_020316639 | hypothetical protein | - |
| HV105_RS30030 (HV105_30030) | 192779..192997 | + | 219 | WP_004171484 | hypothetical protein | - |
| HV105_RS30035 (HV105_30035) | 192998..193315 | + | 318 | WP_020316633 | hypothetical protein | - |
| HV105_RS30040 (HV105_30040) | 193382..193786 | + | 405 | WP_032419926 | hypothetical protein | - |
| HV105_RS30045 (HV105_30045) | 193783..194073 | + | 291 | WP_032419928 | hypothetical protein | - |
| HV105_RS30050 (HV105_30050) | 194081..194479 | + | 399 | WP_020316629 | hypothetical protein | - |
| HV105_RS30055 (HV105_30055) | 194551..197190 | + | 2640 | WP_020316635 | type IV secretion system protein TraC | virb4 |
| HV105_RS30060 (HV105_30060) | 197190..197579 | + | 390 | WP_020316628 | type-F conjugative transfer system protein TrbI | - |
| HV105_RS30065 (HV105_30065) | 197579..198205 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| HV105_RS30070 (HV105_30070) | 198249..199208 | + | 960 | WP_202386622 | conjugal transfer pilus assembly protein TraU | traU |
| HV105_RS30075 (HV105_30075) | 199221..199859 | + | 639 | WP_020316653 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV105_RS30080 (HV105_30080) | 199918..201873 | + | 1956 | WP_020316657 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV105_RS30085 (HV105_30085) | 201906..202187 | + | 282 | WP_032419932 | hypothetical protein | - |
| HV105_RS30090 (HV105_30090) | 202177..202404 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| HV105_RS30095 (HV105_30095) | 202415..202741 | + | 327 | WP_020316638 | hypothetical protein | - |
| HV105_RS30100 (HV105_30100) | 202762..203514 | + | 753 | WP_020316647 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV105_RS30105 (HV105_30105) | 203525..203764 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV105_RS30110 (HV105_30110) | 203736..204293 | + | 558 | WP_020804677 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV105_RS30115 (HV105_30115) | 204339..204782 | + | 444 | WP_020804675 | F-type conjugal transfer protein TrbF | - |
| HV105_RS30120 (HV105_30120) | 204760..206139 | + | 1380 | WP_072198238 | conjugal transfer pilus assembly protein TraH | traH |
| HV105_RS30125 (HV105_30125) | 206139..208961 | + | 2823 | WP_020316632 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV105_RS30130 (HV105_30130) | 208985..209587 | + | 603 | WP_048263624 | hypothetical protein | - |
| HV105_RS30135 (HV105_30135) | 209838..210569 | + | 732 | WP_016831056 | conjugal transfer complement resistance protein TraT | - |
| HV105_RS30140 (HV105_30140) | 210929..213220 | + | 2292 | WP_020316650 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5692 | GenBank | NZ_CP058119 |
| Plasmid name | pRHBSTW-00167_2 | Incompatibility group | IncR |
| Plasmid size | 223875 bp | Coordinate of oriT [Strand] | 185268..185316 [-] |
| Host baterium | Klebsiella michiganensis strain RHBSTW-00167 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merD, merA, merC, merP, merT, merR, arsH, arsC, arsB |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |