Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105220 |
| Name | oriT_RHBSTW-00165|unnamed |
| Organism | Klebsiella grimontii strain RHBSTW-00165 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055365 (41276..41324 [+], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_RHBSTW-00165|unnamed
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file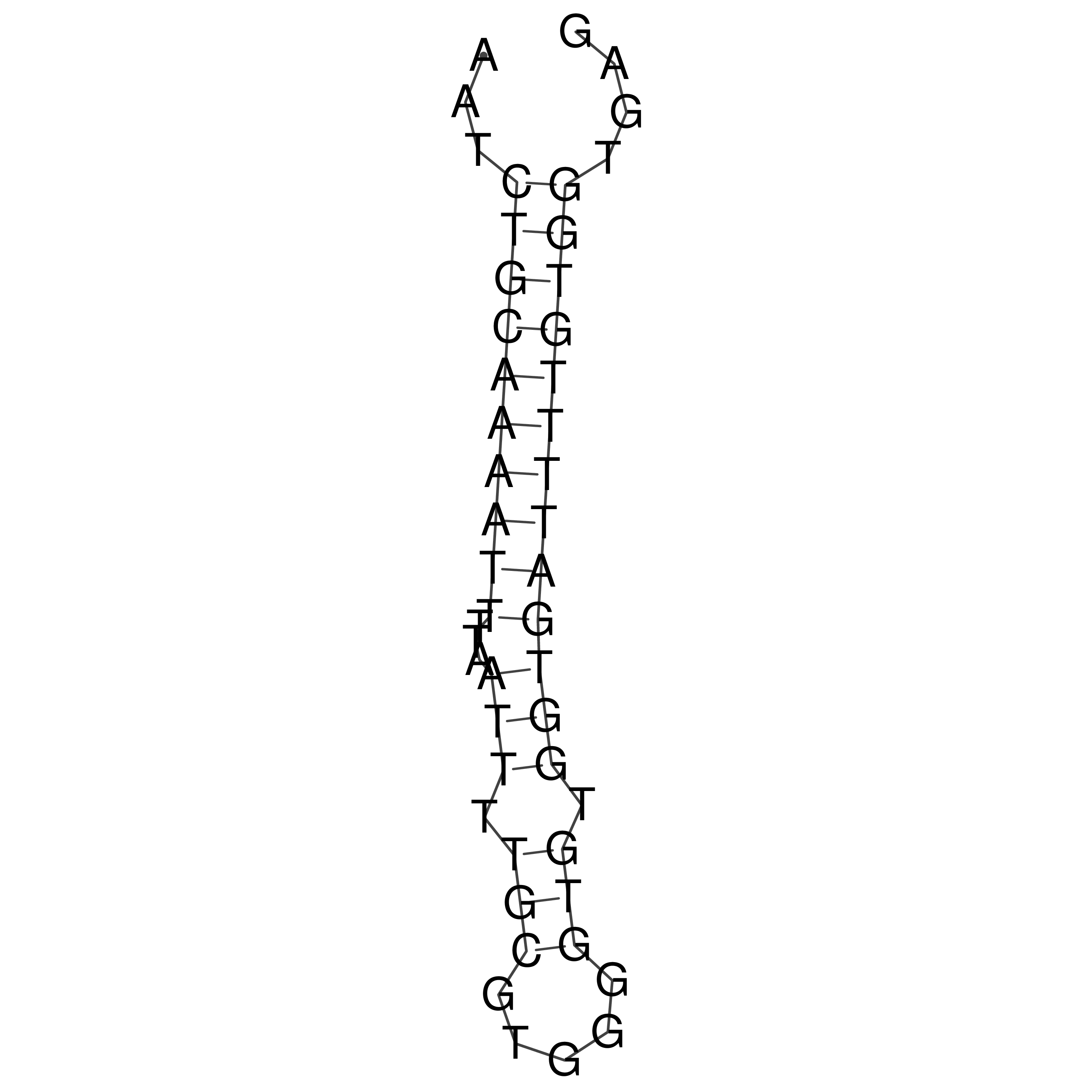
Relaxase
| ID | 3591 | GenBank | WP_181481502 |
| Name | traI_HV104_RS28140_RHBSTW-00165|unnamed |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190442.61 Da Isoelectric Point: 5.6407
>WP_181481502.1 conjugative transfer relaxase/helicase TraI [Klebsiella grimontii]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYASQIAFGKLYREMLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPSEKMVEWVNT
LKETGFDIHSYREDADKRAAEIARAPVSPVKTDGPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLEA
RPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRHAPF
SDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLAGDARLAGETVTG
KSALQDGTAFIPGGSLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGNQTTADIISEPDKNVRYSRLAQEFAVSVREGQESVAQISGTREQNALNNVIRDTLKNEGVLGAKEI
TVTALTPVWLDSKSRGVRDYYREGMVMERWDSETRKHDRFVIDRVTASSNMLTLRGKEGERLDLKVSAVD
SQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFVSVTQRELDNATLNQLAQSGSHIRLYSAQDVARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIASQKAGLHTPAEQAIHLSIPLLESENLTFTRPQLLATALETGGGKVPMTEIESTIR
RQIKAGSLLNVPVAPGQGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMADLTSGQRAATRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAINLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLTDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDINSALTTIEQVTPAQVPRQDGAWVPDNSVVEFTPAQEEAIQK
ALNKGESVPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNDDRRALNSLIHDARRDNGETGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLTDSEGKERLISPREASAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTMDPKADEAQRHIDLAYAITA
HGAQGASEPYAIALEGVAGGRERMASFESAYVGLSRMKQHVQVYTDSREGWIKAIRNSPEKATAHDILEP
RNDRAVKNAGQLFGRAQPLDETAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDKDGRLESIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEALTGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQHREMERQTEQATREVSGEEKKAGEPG
EQIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQESAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLDGKVDKQVFTDLLKGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSVMAMIGGDKRLIDAHNQAVTEAVKQVETLAATRVMTDGKSETVLTGNLI
VAKFNHDTNRNQEPQLHTHAVVINATQNGDKWQSLGTDTVGKTGFIENVYASQIAFGKLYREMLKPLVEK
LGYETEVTGKHGMWEMKDVPVEPFSTRSQEVREAAGPDASLKSRDVATLDTRKSKETLDPSEKMVEWVNT
LKETGFDIHSYREDADKRAAEIARAPVSPVKTDGPDISDVVTKAIAGLSDRKVQFTYADLLARTVSQLEA
RPGVFEQARTGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVSVTPDASVVRHAPF
SDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHIIAADNRSREFLAGDARLAGETVTG
KSALQDGTAFIPGGSLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGNQTTADIISEPDKNVRYSRLAQEFAVSVREGQESVAQISGTREQNALNNVIRDTLKNEGVLGAKEI
TVTALTPVWLDSKSRGVRDYYREGMVMERWDSETRKHDRFVIDRVTASSNMLTLRGKEGERLDLKVSAVD
SQWTLFRADKLPVAEGERLAVLGKIPDTRLKGGESVTVLKAEAGQLTVQRPGQKTTQSLSVGSSPFDGIK
VGHGWVESPGRSVSETATVFVSVTQRELDNATLNQLAQSGSHIRLYSAQDVARTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIASQKAGLHTPAEQAIHLSIPLLESENLTFTRPQLLATALETGGGKVPMTEIESTIR
RQIKAGSLLNVPVAPGQGNDLLISRQTWDAEKSILTKVLEGKEAVAPLMDSVPGSLMADLTSGQRAATRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAINLLPEETRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLTDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDINSALTTIEQVTPAQVPRQDGAWVPDNSVVEFTPAQEEAIQK
ALNKGESVPAGQPATLYEALVKDYTGRTPEAQSQTLIITHLNDDRRALNSLIHDARRDNGETGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLTDSEGKERLISPREASAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKSVSGDSVTLSDGKTTRTMDPKADEAQRHIDLAYAITA
HGAQGASEPYAIALEGVAGGRERMASFESAYVGLSRMKQHVQVYTDSREGWIKAIRNSPEKATAHDILEP
RNDRAVKNAGQLFGRAQPLDETAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKDAGIW
LSPLTDKDGRLESIGGEGRIMGDETARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEALTGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQHREMERQTEQATREVSGEEKKAGEPG
EQIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQESAVQQVATESLQRDRLQQMERDMVRDLSREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1606 | GenBank | WP_022644939 |
| Name | WP_022644939_RHBSTW-00165|unnamed |
UniProt ID | W8QGP1 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15048.16 Da Isoelectric Point: 4.5248
>WP_022644939.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacteriaceae]
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
MAKVQAYVSDEVAEKINAIVEKRRVEGAKDKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNKAL
LEYVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMLRFFPDEDNEVEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W8QGP1 |
T4CP
| ID | 3551 | GenBank | WP_060415494 |
| Name | traD_HV104_RS28145_RHBSTW-00165|unnamed |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 85951.95 Da Isoelectric Point: 4.9227
>WP_060415494.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Klebsiella/Raoultella group]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTVCVVTFFVASWVLGRQGKQ
QSEDEDTGGRQLSDKPKDVARQIKRDGVASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEREYLKPSEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRMDERLSAQLEAREAEGSHARMLFTPDEVAKDEGDTGNSPEAIKPAGDRIP
TPVNATPSEKTPPAEASGNAPVTPQPATPVMRVTTVPLTRPRPVVPASGATAVAAATDVSAVTSGGTAQE
LAQQPVEADQNMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTINYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTVCVVTFFVASWVLGRQGKQ
QSEDEDTGGRQLSDKPKDVARQIKRDGVASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEREYLKPSEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIQRRLDTRMDERLSAQLEAREAEGSHARMLFTPDEVAKDEGDTGNSPEAIKPAGDRIP
TPVNATPSEKTPPAEASGNAPVTPQPATPVMRVTTVPLTRPRPVVPASGATAVAAATDVSAVTSGGTAQE
LAQQPVEADQNMLPAGMNEDGVIEDMQAYDAWLEGEQIQRDMQRREEVNINHSRSEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14869..41891
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV104_RS28145 (HV104_28155) | 14869..17169 | - | 2301 | WP_060415494 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HV104_RS28150 (HV104_28160) | 17416..18087 | - | 672 | WP_088883382 | hypothetical protein | - |
| HV104_RS28155 (HV104_28165) | 18104..20929 | - | 2826 | WP_060415492 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV104_RS28160 (HV104_28170) | 20929..22299 | - | 1371 | WP_032736846 | conjugal transfer pilus assembly protein TraH | traH |
| HV104_RS28165 (HV104_28175) | 22286..22849 | - | 564 | WP_224250969 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV104_RS28170 (HV104_28180) | 22836..23072 | - | 237 | WP_032736848 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV104_RS28175 (HV104_28185) | 23083..23835 | - | 753 | WP_181481503 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV104_RS28180 (HV104_28190) | 23856..24182 | - | 327 | WP_060415488 | hypothetical protein | - |
| HV104_RS28185 (HV104_28195) | 24228..24470 | - | 243 | WP_060415487 | conjugal transfer protein TrbE | - |
| HV104_RS28190 (HV104_28200) | 24460..25071 | - | 612 | WP_060415486 | hypothetical protein | - |
| HV104_RS28195 (HV104_28205) | 25064..25324 | - | 261 | WP_060415485 | hypothetical protein | - |
| HV104_RS28200 (HV104_28210) | 25356..27311 | - | 1956 | WP_181481504 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV104_RS28205 (HV104_28215) | 27308..27934 | - | 627 | WP_060415483 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV104_RS28210 (HV104_28220) | 27947..28906 | - | 960 | WP_088912317 | conjugal transfer pilus assembly protein TraU | traU |
| HV104_RS28215 (HV104_28225) | 28950..29576 | - | 627 | WP_060415482 | type-F conjugative transfer system protein TraW | traW |
| HV104_RS28220 (HV104_28230) | 29573..29962 | - | 390 | WP_060415481 | type-F conjugative transfer system protein TrbI | - |
| HV104_RS28225 (HV104_28235) | 29962..32601 | - | 2640 | WP_060415480 | type IV secretion system protein TraC | virb4 |
| HV104_RS28230 (HV104_28240) | 32694..33077 | - | 384 | WP_181481505 | hypothetical protein | - |
| HV104_RS28235 (HV104_28245) | 33165..33464 | - | 300 | WP_060415478 | hypothetical protein | - |
| HV104_RS31230 | 33461..33862 | - | 402 | WP_060415477 | hypothetical protein | - |
| HV104_RS28245 (HV104_28255) | 33946..34134 | - | 189 | WP_053390294 | hypothetical protein | - |
| HV104_RS28250 (HV104_28260) | 34141..34419 | - | 279 | WP_060415476 | hypothetical protein | - |
| HV104_RS28255 (HV104_28265) | 34531..35100 | - | 570 | WP_060415475 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV104_RS28260 (HV104_28270) | 35265..35873 | - | 609 | WP_060415474 | conjugal transfer protein TraP | - |
| HV104_RS28265 (HV104_28275) | 35866..37287 | - | 1422 | WP_060415473 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV104_RS28270 (HV104_28280) | 37287..38021 | - | 735 | WP_060415472 | type-F conjugative transfer system secretin TraK | traK |
| HV104_RS28275 (HV104_28285) | 38008..38574 | - | 567 | WP_060415471 | type IV conjugative transfer system protein TraE | traE |
| HV104_RS28280 (HV104_28290) | 38594..38899 | - | 306 | WP_053390196 | type IV conjugative transfer system protein TraL | traL |
| HV104_RS28285 (HV104_28295) | 38913..39281 | - | 369 | WP_060415470 | type IV conjugative transfer system pilin TraA | - |
| HV104_RS28290 (HV104_28300) | 39350..39550 | - | 201 | WP_060415469 | TraY domain-containing protein | - |
| HV104_RS28295 (HV104_28305) | 39635..40336 | - | 702 | WP_022644938 | hypothetical protein | - |
| HV104_RS28300 (HV104_28310) | 40571..40963 | - | 393 | WP_022644939 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV104_RS28305 (HV104_28315) | 41469..41891 | + | 423 | WP_073972322 | transglycosylase SLT domain-containing protein | virB1 |
| HV104_RS28310 (HV104_28320) | 41914..42456 | - | 543 | WP_060415467 | antirestriction protein | - |
| HV104_RS28315 (HV104_28325) | 42451..42819 | + | 369 | WP_080848162 | hypothetical protein | - |
| HV104_RS28320 (HV104_28330) | 43105..43938 | - | 834 | WP_060415466 | N-6 DNA methylase | - |
| HV104_RS28325 (HV104_28335) | 43985..44215 | - | 231 | WP_060415465 | hypothetical protein | - |
| HV104_RS28330 (HV104_28340) | 44304..44651 | - | 348 | WP_064182675 | hypothetical protein | - |
| HV104_RS28335 (HV104_28345) | 44708..45046 | - | 339 | WP_020804855 | hypothetical protein | - |
| HV104_RS28340 (HV104_28350) | 45826..46701 | - | 876 | WP_060415463 | DUF4942 domain-containing protein | - |
Host bacterium
| ID | 5658 | GenBank | NZ_CP055365 |
| Plasmid name | RHBSTW-00165|unnamed | Incompatibility group | IncFII |
| Plasmid size | 123417 bp | Coordinate of oriT [Strand] | 41276..41324 [+] |
| Host baterium | Klebsiella grimontii strain RHBSTW-00165 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |