Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105188 |
| Name | oriT_pRHBSTW-00039_5 |
| Organism | Klebsiella grimontii strain RHBSTW-00039 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP058184 (28821..28869 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_pRHBSTW-00039_5
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file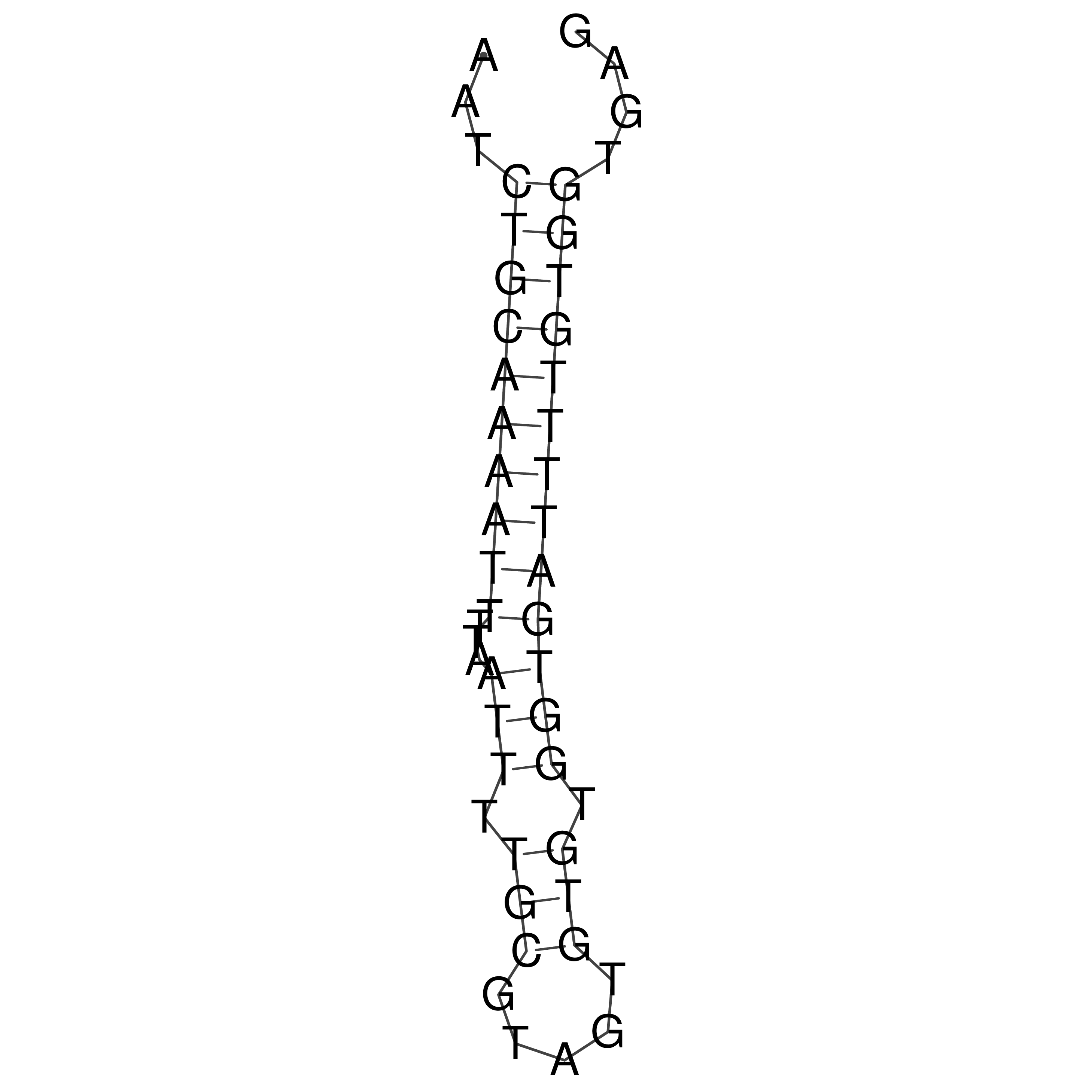
Relaxase
| ID | 3579 | GenBank | WP_181334963 |
| Name | traI_HV042_RS30915_pRHBSTW-00039_5 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190248.05 Da Isoelectric Point: 6.2060
>WP_181334963.1 conjugative transfer relaxase/helicase TraI [Klebsiella grimontii]
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMHHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETMPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLVVGAGVFDGIK
ISHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFAPAQEKAIEK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKRDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSENILANRIALGKIYQNSLRADVES
MGYKTVDAGKNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMHHNVQVLLSDSGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGVRLDLKVSAVD
SQWTLFRAETMPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLVVGAGVFDGIK
ISHGWVESPGRSVSETATVFASLTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPASLMTDLTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIVAGGGRAVSSGDNDQLQPIAPGQPFRLMQQRS
AADIAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGVWAPGSSVVEFAPAQEKAIEK
ALSEGKTLPAGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSDGKERYISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVAGDSVTLSDGKTTRTLTPKADQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDSREGWIKAIKHSPEKATAHDILEP
RNDRAVKSADLLFGRARPLDETAAGRAALQQSGLAQGNSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNDAARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPV
DRVKEVIGDVIRGLERDRPGPEKTTLPDDPQFRRQEAAIQQVASESLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1599 | GenBank | WP_032426796 |
| Name | WP_032426796_pRHBSTW-00039_5 |
UniProt ID | _ |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 15036.10 Da Isoelectric Point: 4.5326
>WP_032426796.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Enterobacterales]
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
MGKVQAYPSDEVCEKINAIVEKRRMEGAKEKDVSFSSISTMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESVVKTQFSVNKVLGIECLSPHVKDDPRWQWKGMVQNIQEDVQEVMRTFFPDEQEEADE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3542 | GenBank | WP_020316650 |
| Name | traD_HV042_RS30910_pRHBSTW-00039_5 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 85197.99 Da Isoelectric Point: 4.8865
>WP_020316650.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTEQELAQQ
SAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRSLDARVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPAEVTV
SPAPVKATQTTKMPAEEPSVRATEPPVLRVTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTEQELAQQ
SAEQGQDMLPAGMNEDGVIEDMQAYDAWADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 31290..56739
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV042_RS30715 (HV042_30720) | 26469..26816 | + | 348 | WP_022644944 | hypothetical protein | - |
| HV042_RS30720 (HV042_30725) | 26909..27055 | + | 147 | WP_022644943 | hypothetical protein | - |
| HV042_RS31385 | 27103..27339 | + | 237 | Protein_39 | SAM-dependent DNA methyltransferase | - |
| HV042_RS30730 (HV042_30735) | 27688..28230 | + | 543 | WP_022644941 | antirestriction protein | - |
| HV042_RS30735 (HV042_30740) | 28254..28676 | - | 423 | WP_072205880 | transglycosylase SLT domain-containing protein | - |
| HV042_RS30740 (HV042_30745) | 29182..29574 | + | 393 | WP_032426796 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV042_RS30745 (HV042_30750) | 29812..30504 | + | 693 | WP_032426795 | conjugal transfer protein TrbJ | - |
| HV042_RS30750 (HV042_30755) | 30680..30844 | + | 165 | WP_032426810 | TraY domain-containing protein | - |
| HV042_RS30755 (HV042_30760) | 30908..31276 | + | 369 | WP_032426794 | type IV conjugative transfer system pilin TraA | - |
| HV042_RS30760 (HV042_30765) | 31290..31595 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HV042_RS30765 (HV042_30770) | 31615..32181 | + | 567 | WP_020316627 | type IV conjugative transfer system protein TraE | traE |
| HV042_RS30770 (HV042_30775) | 32168..32908 | + | 741 | WP_110222093 | type-F conjugative transfer system secretin TraK | traK |
| HV042_RS30775 (HV042_30780) | 32908..34332 | + | 1425 | WP_181334965 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV042_RS30780 (HV042_30785) | 34325..34921 | + | 597 | WP_103214945 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV042_RS30785 (HV042_30790) | 34944..35513 | + | 570 | WP_023316399 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV042_RS30790 (HV042_30795) | 35645..36055 | + | 411 | WP_032419923 | hypothetical protein | - |
| HV042_RS30795 (HV042_30800) | 36060..36350 | + | 291 | WP_048993668 | hypothetical protein | - |
| HV042_RS30800 (HV042_30805) | 36374..36592 | + | 219 | WP_023316402 | hypothetical protein | - |
| HV042_RS30805 (HV042_30810) | 36593..36904 | + | 312 | WP_023316403 | hypothetical protein | - |
| HV042_RS30810 (HV042_30815) | 36971..37375 | + | 405 | WP_023340941 | hypothetical protein | - |
| HV042_RS30815 (HV042_30820) | 37372..37662 | + | 291 | WP_023340940 | hypothetical protein | - |
| HV042_RS30820 (HV042_30825) | 37670..38068 | + | 399 | WP_074186017 | hypothetical protein | - |
| HV042_RS30825 (HV042_30830) | 38140..40779 | + | 2640 | WP_181334960 | type IV secretion system protein TraC | virb4 |
| HV042_RS30830 (HV042_30835) | 40779..41168 | + | 390 | WP_020314627 | type-F conjugative transfer system protein TrbI | - |
| HV042_RS30835 (HV042_30840) | 41168..41794 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| HV042_RS30840 (HV042_30845) | 41838..42797 | + | 960 | WP_239689303 | conjugal transfer pilus assembly protein TraU | traU |
| HV042_RS30845 (HV042_30850) | 42810..43448 | + | 639 | WP_032456788 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV042_RS30850 (HV042_30855) | 43507..45462 | + | 1956 | WP_181334961 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV042_RS30855 (HV042_30860) | 45494..45748 | + | 255 | WP_004195500 | conjugal transfer protein TrbE | - |
| HV042_RS30860 (HV042_30865) | 45726..45974 | + | 249 | WP_004152675 | hypothetical protein | - |
| HV042_RS30865 (HV042_30870) | 45987..46313 | + | 327 | WP_004194451 | hypothetical protein | - |
| HV042_RS30870 (HV042_30875) | 46334..47086 | + | 753 | WP_009309874 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV042_RS30875 (HV042_30880) | 47097..47336 | + | 240 | WP_009309875 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV042_RS30880 (HV042_30885) | 47308..47880 | + | 573 | WP_009309876 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV042_RS30885 (HV042_30890) | 47873..48301 | + | 429 | WP_004152689 | hypothetical protein | - |
| HV042_RS30890 (HV042_30895) | 48288..49658 | + | 1371 | WP_009309877 | conjugal transfer pilus assembly protein TraH | traH |
| HV042_RS30895 (HV042_30900) | 49658..52480 | + | 2823 | WP_181334962 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV042_RS30900 (HV042_30905) | 52504..53106 | + | 603 | WP_048263624 | hypothetical protein | - |
| HV042_RS30905 (HV042_30910) | 53357..54088 | + | 732 | WP_016831056 | conjugal transfer complement resistance protein TraT | - |
| HV042_RS30910 (HV042_30915) | 54448..56739 | + | 2292 | WP_020316650 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5626 | GenBank | NZ_CP058184 |
| Plasmid name | pRHBSTW-00039_5 | Incompatibility group | - |
| Plasmid size | 67313 bp | Coordinate of oriT [Strand] | 28821..28869 [-] |
| Host baterium | Klebsiella grimontii strain RHBSTW-00039 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |