Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105182 |
| Name | oriT_pRHBSTW-00577_2 |
| Organism | Klebsiella grimontii strain RHBSTW-00577 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP056702 (114611..114660 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pRHBSTW-00577_2
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file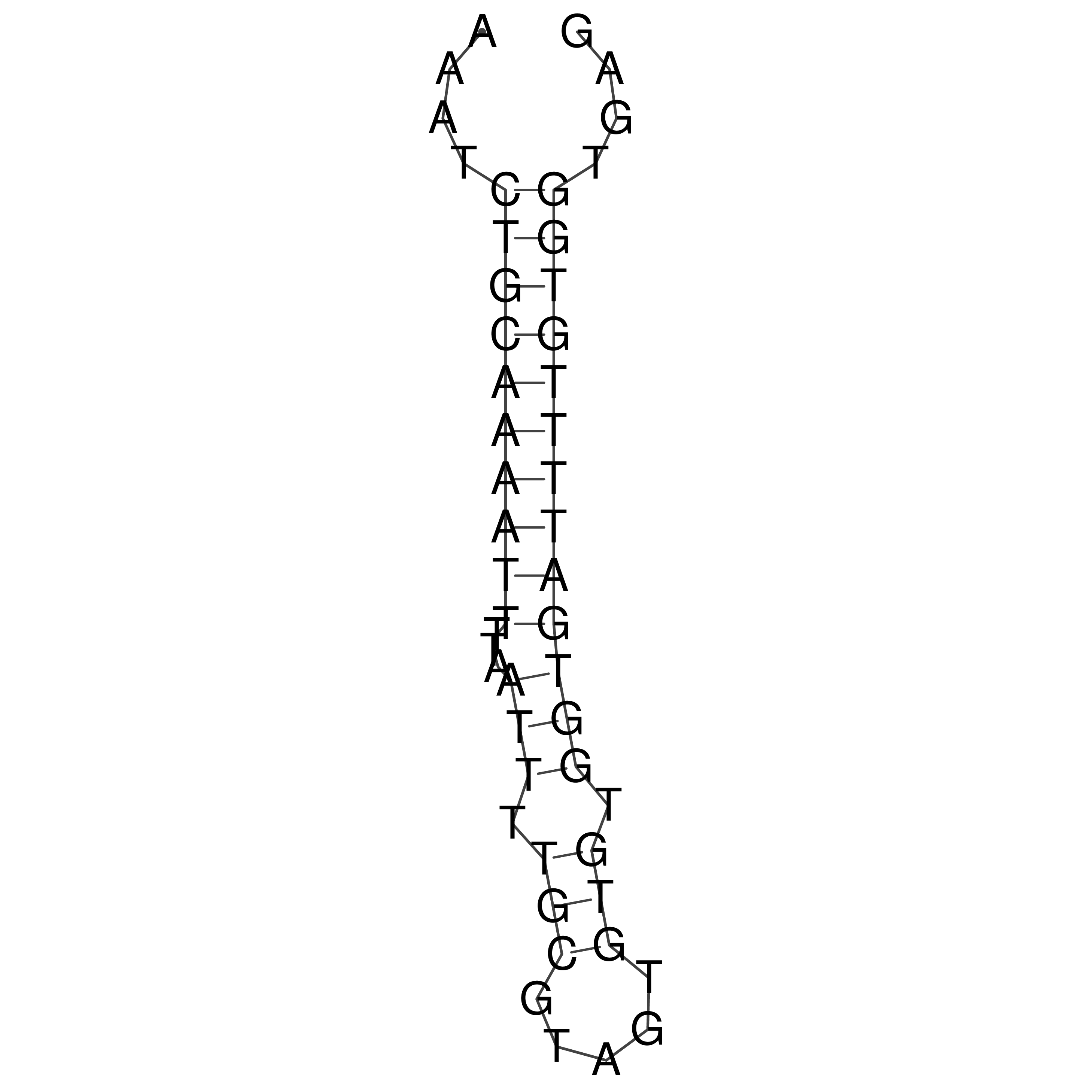
Relaxase
| ID | 3574 | GenBank | WP_048270266 |
| Name | traI_HV240_RS27815_pRHBSTW-00577_2 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190566.21 Da Isoelectric Point: 5.9711
>WP_048270266.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDGGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDGGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEDGQLTVQRPGQKTTQTLAVGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEEAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3537 | GenBank | WP_013214038 |
| Name | traD_HV240_RS27820_pRHBSTW-00577_2 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 86010.07 Da Isoelectric Point: 5.1787
>WP_013214038.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPEPVKAPPTIKRPAAEPSVRTTEPSVLRVTTVPLIKPKAAAAAAAASTASSSGAPATAAGGTQ
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 85571..115218
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV240_RS27820 (HV240_27820) | 85571..87883 | - | 2313 | WP_013214038 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HV240_RS27825 (HV240_27825) | 88010..88699 | - | 690 | WP_162497697 | hypothetical protein | - |
| HV240_RS27830 (HV240_27830) | 88892..89623 | - | 732 | WP_004152629 | conjugal transfer complement resistance protein TraT | - |
| HV240_RS27835 (HV240_27835) | 89932..90456 | - | 525 | WP_040206919 | hypothetical protein | - |
| HV240_RS27840 (HV240_27840) | 90467..93310 | - | 2844 | WP_094833276 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV240_RS27845 (HV240_27845) | 93310..94689 | - | 1380 | WP_181512256 | conjugal transfer pilus assembly protein TraH | traH |
| HV240_RS27850 (HV240_27850) | 94667..95110 | - | 444 | WP_040207148 | F-type conjugal transfer protein TrbF | - |
| HV240_RS27855 (HV240_27855) | 95156..95713 | - | 558 | WP_032445241 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV240_RS27860 (HV240_27860) | 95685..95924 | - | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV240_RS27865 (HV240_27865) | 95935..96687 | - | 753 | WP_032429773 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV240_RS27870 (HV240_27870) | 96708..97034 | - | 327 | WP_016529362 | hypothetical protein | - |
| HV240_RS30015 | 97080..97265 | - | 186 | WP_032445239 | hypothetical protein | - |
| HV240_RS27875 (HV240_27875) | 97262..97498 | - | 237 | WP_032445238 | conjugal transfer protein TrbE | - |
| HV240_RS27880 (HV240_27880) | 97530..99485 | - | 1956 | WP_140599676 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV240_RS27885 (HV240_27885) | 99544..100182 | - | 639 | WP_040207142 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV240_RS27890 (HV240_27890) | 100195..101184 | - | 990 | WP_009309872 | conjugal transfer pilus assembly protein TraU | traU |
| HV240_RS27895 (HV240_27895) | 101181..101570 | - | 390 | WP_004144408 | hypothetical protein | - |
| HV240_RS27900 (HV240_27900) | 101612..102238 | - | 627 | WP_032445234 | type-F conjugative transfer system protein TraW | traW |
| HV240_RS27905 (HV240_27905) | 102238..102627 | - | 390 | WP_032445233 | type-F conjugative transfer system protein TrbI | - |
| HV240_RS27910 (HV240_27910) | 102627..105266 | - | 2640 | WP_032431388 | type IV secretion system protein TraC | virb4 |
| HV240_RS27915 (HV240_27915) | 105338..105736 | - | 399 | WP_181512257 | hypothetical protein | - |
| HV240_RS27920 (HV240_27920) | 105744..106043 | - | 300 | WP_055322554 | hypothetical protein | - |
| HV240_RS27925 (HV240_27925) | 106086..106490 | - | 405 | WP_181512258 | hypothetical protein | - |
| HV240_RS27930 (HV240_27930) | 106557..106874 | - | 318 | WP_181512259 | hypothetical protein | - |
| HV240_RS27935 (HV240_27935) | 106875..107093 | - | 219 | WP_004195468 | hypothetical protein | - |
| HV240_RS30020 (HV240_27940) | 107117..107381 | - | 265 | Protein_108 | hypothetical protein | - |
| HV240_RS27940 (HV240_27945) | 107986..108570 | - | 585 | WP_181512260 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV240_RS27945 (HV240_27950) | 108593..109189 | - | 597 | WP_181512261 | conjugal transfer pilus-stabilizing protein TraP | - |
| HV240_RS27950 (HV240_27955) | 109182..110606 | - | 1425 | WP_004233801 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV240_RS27955 (HV240_27960) | 110606..111346 | - | 741 | WP_029498376 | type-F conjugative transfer system secretin TraK | traK |
| HV240_RS27960 (HV240_27965) | 111333..111899 | - | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| HV240_RS27965 (HV240_27970) | 111919..112224 | - | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HV240_RS27970 (HV240_27975) | 112238..112606 | - | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| HV240_RS27975 (HV240_27980) | 112668..112874 | - | 207 | WP_171773970 | TraY domain-containing protein | - |
| HV240_RS27980 (HV240_27985) | 113007..113726 | - | 720 | WP_014386192 | conjugal transfer protein | - |
| HV240_RS27985 (HV240_27990) | 113926..114342 | - | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV240_RS27990 (HV240_27995) | 114733..115218 | + | 486 | WP_001568108 | transglycosylase SLT domain-containing protein | virB1 |
| HV240_RS27995 (HV240_28000) | 115251..115580 | - | 330 | WP_011977736 | DUF5983 family protein | - |
| HV240_RS28000 (HV240_28005) | 115613..116434 | - | 822 | WP_023316395 | DUF932 domain-containing protein | - |
| HV240_RS28005 (HV240_28010) | 117356..118645 | - | 1290 | WP_004574636 | ISL3-like element ISPpu12 family transposase | - |
| HV240_RS28010 (HV240_28015) | 118667..119179 | - | 513 | WP_004863699 | signal peptidase II | - |
| HV240_RS28015 (HV240_28020) | 119183..120079 | - | 897 | WP_004574643 | cation transporter | - |
Host bacterium
| ID | 5620 | GenBank | NZ_CP056702 |
| Plasmid name | pRHBSTW-00577_2 | Incompatibility group | IncFII |
| Plasmid size | 144021 bp | Coordinate of oriT [Strand] | 114611..114660 [+] |
| Host baterium | Klebsiella grimontii strain RHBSTW-00577 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | merR, merT, merP, merC, merA, merD, merE |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |