Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105170 |
| Name | oriT_pRHBSTW-00814_2 |
| Organism | Escherichia marmotae strain RHBSTW-00814 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP056160 (48897..48980 [+], 84 nt) |
| oriT length | 84 nt |
| IRs (inverted repeats) | 28..33, 37..42 (GCCATC..GATGGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 84 nt
>oriT_pRHBSTW-00814_2
ACGGGACAGGATATGTTTTGTTGAACCGCCATCTTCGATGGCCGTTCAACAAAACCCCTGGTCAGGGCGCAGCCCCGACACCCC
ACGGGACAGGATATGTTTTGTTGAACCGCCATCTTCGATGGCCGTTCAACAAAACCCCTGGTCAGGGCGCAGCCCCGACACCCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file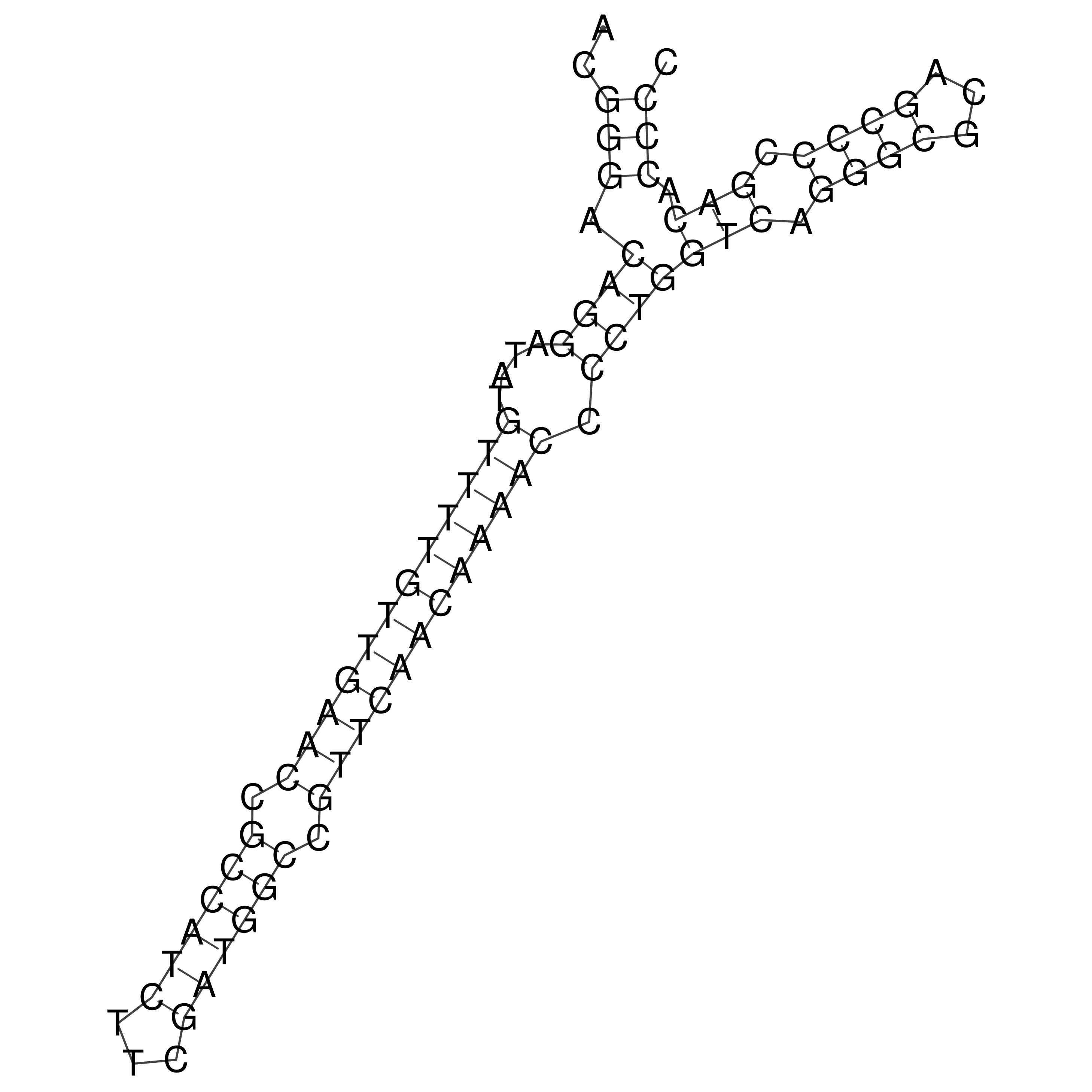
Relaxase
| ID | 3568 | GenBank | WP_181475202 |
| Name | Relaxase_HV284_RS24330_pRHBSTW-00814_2 |
UniProt ID | _ |
| Length | 644 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 644 a.a. Molecular weight: 74898.88 Da Isoelectric Point: 6.6215
>WP_181475202.1 TraI/MobA(P) family conjugative relaxase [Escherichia marmotae]
MDKKPGELLSPDNPYVRPVRSREAIVEHLGQFIEEGMDLYRNHVLSRSDDGRQQVLCGEVLCETNCFGIE
TAAAEMDAVASQNRRCQDPVYHAILSWREEDNPTNEQIFECARYALKALNMSDHQYVFAIHRDTEHVHCH
ITVNRINPKTYRAASLYDDYFRLHKACRELEQKYGFTPDNGCCQKDENGNWIRKQEEFKSIPRKARQMEY
HADKESLYSYAVDEARHTIGKILHSGEATWEALHTELIRLGLELKEKGEGLAVYSRHDDTVTPIKASTLH
PDLTKRVLEPYLGKFTPSPVVENYFDENDNYLGSNYPQEYVYDVRAHKRDIGASSARREERADAREALKA
RYNAYRAGWTRPEMDRDEIRARFYHVSGQFAWRRAQVRAEIRDPLLRKLAYNVLALERRQAMIALKETLR
EERKAFYARPENRRMGYWQWVEQQALNNDAAAISCLRGRFYMQKKMQQENRLSENAIVCAVADDIRPYEI
EGYQTQITRDGRIQYCQDGEVKLQDTGDRIEIVDPYSQDGQHIAGAMVLAEVKSGERMYFAGEPQFVNKA
CSIVPWFNEGSEKPLPLTDRAQRQMAGYDTAPASPQKDEPEIARESLRWLMEGAPKTPVQEDVRQQQKHD
YVHDSSYSSRYRRH
MDKKPGELLSPDNPYVRPVRSREAIVEHLGQFIEEGMDLYRNHVLSRSDDGRQQVLCGEVLCETNCFGIE
TAAAEMDAVASQNRRCQDPVYHAILSWREEDNPTNEQIFECARYALKALNMSDHQYVFAIHRDTEHVHCH
ITVNRINPKTYRAASLYDDYFRLHKACRELEQKYGFTPDNGCCQKDENGNWIRKQEEFKSIPRKARQMEY
HADKESLYSYAVDEARHTIGKILHSGEATWEALHTELIRLGLELKEKGEGLAVYSRHDDTVTPIKASTLH
PDLTKRVLEPYLGKFTPSPVVENYFDENDNYLGSNYPQEYVYDVRAHKRDIGASSARREERADAREALKA
RYNAYRAGWTRPEMDRDEIRARFYHVSGQFAWRRAQVRAEIRDPLLRKLAYNVLALERRQAMIALKETLR
EERKAFYARPENRRMGYWQWVEQQALNNDAAAISCLRGRFYMQKKMQQENRLSENAIVCAVADDIRPYEI
EGYQTQITRDGRIQYCQDGEVKLQDTGDRIEIVDPYSQDGQHIAGAMVLAEVKSGERMYFAGEPQFVNKA
CSIVPWFNEGSEKPLPLTDRAQRQMAGYDTAPASPQKDEPEIARESLRWLMEGAPKTPVQEDVRQQQKHD
YVHDSSYSSRYRRH
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3532 | GenBank | WP_249415932 |
| Name | t4cp2_HV284_RS24520_pRHBSTW-00814_2 |
UniProt ID | _ |
| Length | 494 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 494 a.a. Molecular weight: 55978.92 Da Isoelectric Point: 6.5712
>WP_249415932.1 hypothetical protein [Escherichia marmotae]
MLPKVDGSSADWQDKARPMLYGLVYSLHYKCRRENLRFSLAMLHKHLPLKEMAALYIEGKQNGWHQEGIN
ALENYLSTLAGFQMDLVSRPGEWDQGVFDQHGFLIQQFSKMLAMFNDVYGHIFANDAGDIDLRDVLHNDR
ALVVLIPALELSKSESASLGKLYISAKRMVIARDLGYHLEGKREEVLTAKKYRNRFPFPDIHDELGSYFA
PGMDDLAAQMRSLGYMLIISAQDIQRFIAQFRGEYQTVNANLLVKWFMALQDEKDTYELARSTAGKDYFA
ELGEVKRSPGLFGNQFEDATTTHIREKDRISLDELKNLNPGEGFISFKSALVPCNAVYIPDDEKLSSKLQ
MRINRFIDIGRPSESELVKKNPSLARLLPPDSQEVSLVLQRTDEIMAGMAPRNPPPVVDPILARLAEITL
NLDNRCDISYTPEQRGILLFEAAREAMKHSRGRWFLAQTRHRPISVSDQVAHKLATDSTAFPSPVNTDKG
TDPQ
MLPKVDGSSADWQDKARPMLYGLVYSLHYKCRRENLRFSLAMLHKHLPLKEMAALYIEGKQNGWHQEGIN
ALENYLSTLAGFQMDLVSRPGEWDQGVFDQHGFLIQQFSKMLAMFNDVYGHIFANDAGDIDLRDVLHNDR
ALVVLIPALELSKSESASLGKLYISAKRMVIARDLGYHLEGKREEVLTAKKYRNRFPFPDIHDELGSYFA
PGMDDLAAQMRSLGYMLIISAQDIQRFIAQFRGEYQTVNANLLVKWFMALQDEKDTYELARSTAGKDYFA
ELGEVKRSPGLFGNQFEDATTTHIREKDRISLDELKNLNPGEGFISFKSALVPCNAVYIPDDEKLSSKLQ
MRINRFIDIGRPSESELVKKNPSLARLLPPDSQEVSLVLQRTDEIMAGMAPRNPPPVVDPILARLAEITL
NLDNRCDISYTPEQRGILLFEAAREAMKHSRGRWFLAQTRHRPISVSDQVAHKLATDSTAFPSPVNTDKG
TDPQ
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 85820..113587
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV284_RS24515 (HV284_24525) | 83084..83584 | - | 501 | WP_181475210 | phospholipase D family protein | - |
| HV284_RS24520 (HV284_24530) | 83614..85098 | - | 1485 | WP_249415932 | hypothetical protein | - |
| HV284_RS25320 | 85273..85830 | - | 558 | WP_249415933 | hypothetical protein | - |
| HV284_RS24525 (HV284_24535) | 85820..86740 | - | 921 | WP_181475168 | hypothetical protein | trbB |
| HV284_RS24530 (HV284_24540) | 86750..88123 | - | 1374 | WP_181475169 | conjugal transfer protein TrbA | trbA |
| HV284_RS24535 (HV284_24545) | 88410..88547 | - | 138 | WP_181475170 | Hok/Gef family protein | - |
| HV284_RS24540 (HV284_24550) | 88907..89560 | - | 654 | WP_181475171 | DUF5710 domain-containing protein | - |
| HV284_RS24545 (HV284_24555) | 89599..89955 | - | 357 | WP_107135981 | hypothetical protein | - |
| HV284_RS24550 (HV284_24560) | 90209..90415 | - | 207 | WP_038808981 | hemolysin expression modulator Hha | - |
| HV284_RS24555 (HV284_24565) | 91148..91711 | - | 564 | WP_243053450 | hypothetical protein | - |
| HV284_RS24560 (HV284_24570) | 91724..93877 | - | 2154 | WP_181475172 | DotA/TraY family protein | traY |
| HV284_RS24565 (HV284_24575) | 93961..94305 | - | 345 | WP_215251741 | hypothetical protein | - |
| HV284_RS25325 | 94307..94621 | - | 315 | WP_249415934 | hypothetical protein | - |
| HV284_RS24570 (HV284_24580) | 94957..95250 | - | 294 | WP_063848353 | type II toxin-antitoxin system RelE/ParE family toxin | - |
| HV284_RS24575 (HV284_24585) | 95250..95522 | - | 273 | WP_063848352 | type II toxin-antitoxin system Phd/YefM family antitoxin | - |
| HV284_RS24580 (HV284_24590) | 95659..95805 | - | 147 | WP_181475174 | hypothetical protein | - |
| HV284_RS24585 (HV284_24595) | 95950..96480 | - | 531 | WP_163516558 | conjugal transfer protein TraX | - |
| HV284_RS24590 (HV284_24600) | 96477..97697 | - | 1221 | WP_001039317 | conjugal transfer protein TraW | traW |
| HV284_RS24595 (HV284_24605) | 97721..100777 | - | 3057 | WP_181475175 | ATP-binding protein | traU |
| HV284_RS24600 (HV284_24610) | 100781..101392 | - | 612 | WP_181475176 | hypothetical protein | traT |
| HV284_RS24605 (HV284_24615) | 101410..101655 | - | 246 | WP_181475177 | hypothetical protein | - |
| HV284_RS24610 (HV284_24620) | 101711..102118 | - | 408 | WP_001059622 | DUF6750 family protein | traR |
| HV284_RS24615 (HV284_24625) | 102150..102674 | - | 525 | WP_212768175 | conjugal transfer protein TraQ | traQ |
| HV284_RS24620 (HV284_24630) | 102685..103386 | - | 702 | WP_105289550 | conjugal transfer protein TraP | traP |
| HV284_RS24625 (HV284_24635) | 103387..104508 | - | 1122 | WP_249415935 | conjugal transfer protein TraO | traO |
| HV284_RS24630 (HV284_24640) | 104583..105644 | - | 1062 | WP_181475179 | DotH/IcmK family type IV secretion protein | traN |
| HV284_RS24635 (HV284_24645) | 105641..106312 | - | 672 | WP_097314225 | DotI/IcmL/TraM family protein | traM |
| HV284_RS24640 (HV284_24650) | 106332..106811 | - | 480 | WP_105289547 | hypothetical protein | traL |
| HV284_RS24645 (HV284_24655) | 106826..110437 | - | 3612 | WP_181475180 | DUF5710 domain-containing protein | - |
| HV284_RS24650 (HV284_24660) | 110455..110715 | - | 261 | WP_050939877 | IcmT/TraK family protein | traK |
| HV284_RS24655 (HV284_24665) | 110708..111880 | - | 1173 | WP_176559148 | plasmid transfer ATPase TraJ | virB11 |
| HV284_RS24660 (HV284_24670) | 111886..112668 | - | 783 | WP_181475181 | type IV secretory system conjugative DNA transfer family protein | traI |
| HV284_RS24665 (HV284_24675) | 112665..113090 | - | 426 | WP_181475182 | DotD/TraH family lipoprotein | - |
| HV284_RS24670 (HV284_24680) | 113117..113587 | - | 471 | WP_157917732 | lytic transglycosylase domain-containing protein | virB1 |
| HV284_RS24675 (HV284_24685) | 113587..114084 | - | 498 | WP_181475183 | hypothetical protein | - |
| HV284_RS24680 (HV284_24690) | 114094..114336 | - | 243 | WP_105289541 | hypothetical protein | - |
| HV284_RS24685 (HV284_24695) | 114475..114675 | - | 201 | WP_105289540 | hypothetical protein | - |
| HV284_RS24690 (HV284_24700) | 114665..115342 | - | 678 | WP_181475184 | DUF2913 family protein | - |
| HV284_RS24695 (HV284_24705) | 115440..115952 | - | 513 | WP_105289538 | transcription termination/antitermination NusG family protein | - |
Host bacterium
| ID | 5608 | GenBank | NZ_CP056160 |
| Plasmid name | pRHBSTW-00814_2 | Incompatibility group | IncI1 |
| Plasmid size | 116663 bp | Coordinate of oriT [Strand] | 48897..48980 [+] |
| Host baterium | Escherichia marmotae strain RHBSTW-00814 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | faeC, faeD, faeE, faeF, faeH, faeI, faeJ, papD, papC, papH |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |