Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 105144 |
| Name | oriT_pRHBSTW-00001_2 |
| Organism | Klebsiella pneumoniae strain RHBSTW-00001 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055594 (76153..76202 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pRHBSTW-00001_2
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file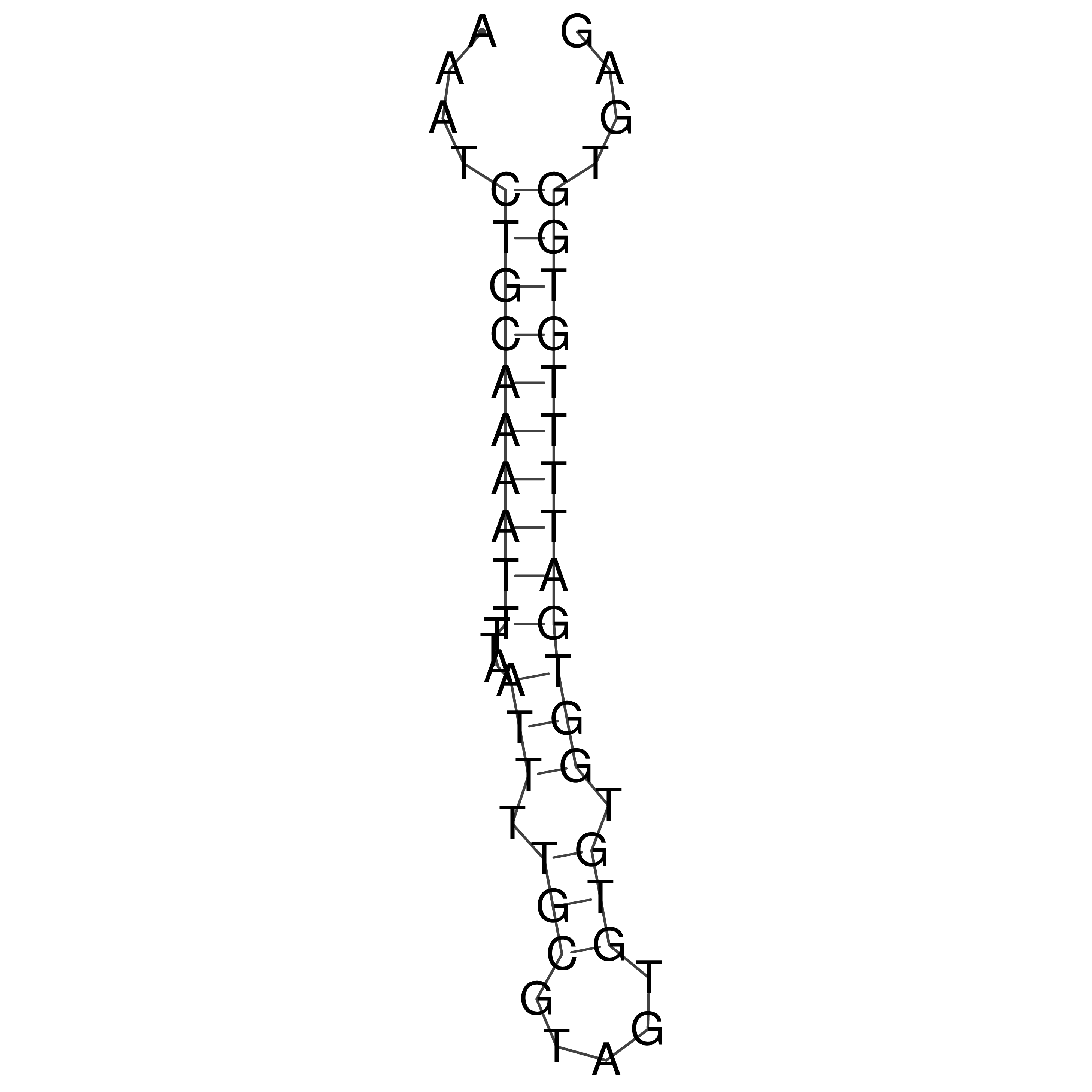
Relaxase
| ID | 3560 | GenBank | WP_151819948 |
| Name | traI_HV026_RS26180_pRHBSTW-00001_2 |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 190451.11 Da Isoelectric Point: 5.9703
>WP_151819948.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacteriaceae]
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDGGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQLELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVIGSMEERWQGKGAELLGLEGKVDKQVFTELLQGKLPDGSDLTRIQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALNQVESLASTRVKKDGVSETVLTGNLI
IARFNHDTSRAQDPQIHTHSVVINATQNGDKWQTLASDTVGKTGFSETILANRIAFGKIYQNSLRADVES
MGYKTVDAGRNGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMNT
LKETGFDIRGYREAADARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLEA
KDGVFDLARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVPF
SDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDGGKRSGTGSALTVLKDSGVNTYR
WQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKDT
TITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDREGDRLDLKVSAVD
SQWTLFRADTLPVAEGERLAVLGKIPDTRLKGGESITVMKVEEGQLTVQRPGQKTTQTLAAGAGVFDGIK
VGHGWVESPGRSVSETATVFASVTQLELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSEQ
LKSRSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTIQ
AQIRSGQLLNVPVAPGRGNDLLISRQAWDAEKSILTRVLEGKDAVAPLMDRVPDSLMTELTAGQRAATRM
ILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVDARTTASFLHD
TQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQRS
AADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIEK
ALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITLP
VLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTLY
RQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAITA
HGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILEP
RNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGIW
LSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDRP
WNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEPA
DRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3525 | GenBank | WP_040215031 |
| Name | traD_HV026_RS26175_pRHBSTW-00001_2 |
UniProt ID | _ |
| Length | 760 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 760 a.a. Molecular weight: 85020.03 Da Isoelectric Point: 4.9634
>WP_040215031.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLTMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDERVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTSFVFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLTMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSREIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDERVDARLSALLEAREAEGSLARALFTPDAPEPGPADTDSHAGEQPEPVSPPA
PAEMTVSPAPVKAPPTTKRPAAEPPVLPVTPIPLLKPKAAAAAATASSAGTPAAAAGGTEQELAQQSAEQ
GQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 75595..105595
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV026_RS25960 (HV026_25975) | 70693..71043 | + | 351 | WP_004153414 | hypothetical protein | - |
| HV026_RS25965 (HV026_25980) | 71677..72030 | + | 354 | WP_004152748 | hypothetical protein | - |
| HV026_RS25970 (HV026_25985) | 72087..72434 | + | 348 | WP_004152749 | hypothetical protein | - |
| HV026_RS25975 (HV026_25990) | 72529..72675 | + | 147 | WP_004152750 | hypothetical protein | - |
| HV026_RS25980 (HV026_25995) | 72726..73559 | + | 834 | WP_004152751 | N-6 DNA methylase | - |
| HV026_RS25985 (HV026_26000) | 74379..75200 | + | 822 | WP_004152492 | DUF932 domain-containing protein | - |
| HV026_RS25990 (HV026_26005) | 75233..75562 | + | 330 | WP_011977736 | DUF5983 family protein | - |
| HV026_RS25995 (HV026_26010) | 75595..76080 | - | 486 | WP_022631514 | transglycosylase SLT domain-containing protein | virB1 |
| HV026_RS26000 (HV026_26015) | 76471..76887 | + | 417 | WP_072145360 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV026_RS26005 (HV026_26020) | 77087..77806 | + | 720 | WP_015065526 | hypothetical protein | - |
| HV026_RS26010 (HV026_26025) | 77939..78142 | + | 204 | WP_171486139 | TraY domain-containing protein | - |
| HV026_RS26015 (HV026_26030) | 78207..78575 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| HV026_RS26020 (HV026_26035) | 78589..78894 | + | 306 | WP_004144424 | type IV conjugative transfer system protein TraL | traL |
| HV026_RS26025 (HV026_26040) | 78914..79480 | + | 567 | WP_004144423 | type IV conjugative transfer system protein TraE | traE |
| HV026_RS26030 (HV026_26045) | 79467..80207 | + | 741 | WP_004152497 | type-F conjugative transfer system secretin TraK | traK |
| HV026_RS26035 (HV026_26050) | 80207..81631 | + | 1425 | WP_032426793 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV026_RS26040 (HV026_26055) | 81745..82329 | + | 585 | WP_004161368 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV026_RS26045 (HV026_26060) | 82461..82871 | + | 411 | WP_004152499 | hypothetical protein | - |
| HV026_RS26050 (HV026_26065) | 82977..83195 | + | 219 | WP_004152501 | hypothetical protein | - |
| HV026_RS26055 (HV026_26070) | 83196..83507 | + | 312 | WP_004152502 | hypothetical protein | - |
| HV026_RS26060 (HV026_26075) | 83574..83978 | + | 405 | WP_004152503 | hypothetical protein | - |
| HV026_RS26065 (HV026_26080) | 84021..84410 | + | 390 | WP_004153076 | hypothetical protein | - |
| HV026_RS26070 (HV026_26085) | 84418..84816 | + | 399 | WP_023179972 | hypothetical protein | - |
| HV026_RS26075 (HV026_26090) | 84888..87527 | + | 2640 | WP_004152505 | type IV secretion system protein TraC | virb4 |
| HV026_RS26080 (HV026_26095) | 87527..87916 | + | 390 | WP_004152506 | type-F conjugative transfer system protein TrbI | - |
| HV026_RS26085 (HV026_26100) | 87916..88542 | + | 627 | WP_181467186 | type-F conjugative transfer system protein TraW | traW |
| HV026_RS26090 (HV026_26105) | 88584..88973 | + | 390 | WP_032445235 | hypothetical protein | - |
| HV026_RS26095 (HV026_26110) | 88970..89959 | + | 990 | WP_011977785 | conjugal transfer pilus assembly protein TraU | traU |
| HV026_RS26100 (HV026_26115) | 89972..90610 | + | 639 | WP_004193871 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV026_RS26105 (HV026_26120) | 90669..92624 | + | 1956 | WP_030003482 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV026_RS26110 (HV026_26125) | 92656..92910 | + | 255 | WP_004152674 | conjugal transfer protein TrbE | - |
| HV026_RS26115 (HV026_26130) | 92888..93136 | + | 249 | WP_004152675 | hypothetical protein | - |
| HV026_RS26120 (HV026_26135) | 93149..93475 | + | 327 | WP_004144402 | hypothetical protein | - |
| HV026_RS26125 (HV026_26140) | 93496..94248 | + | 753 | WP_020803149 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV026_RS26130 (HV026_26145) | 94259..94498 | + | 240 | WP_004152687 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV026_RS26135 (HV026_26150) | 94470..95042 | + | 573 | WP_023292147 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV026_RS26140 (HV026_26155) | 95035..95463 | + | 429 | WP_015065560 | hypothetical protein | - |
| HV026_RS26145 (HV026_26160) | 95450..96820 | + | 1371 | WP_004159274 | conjugal transfer pilus assembly protein TraH | traH |
| HV026_RS26150 (HV026_26165) | 96820..99663 | + | 2844 | WP_032420975 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV026_RS26155 (HV026_26170) | 99674..100198 | + | 525 | WP_015065538 | hypothetical protein | - |
| HV026_RS26160 (HV026_26175) | 100527..101495 | - | 969 | WP_072193976 | IS5-like element IS903B family transposase | - |
| HV026_RS26165 (HV026_26180) | 101573..102304 | + | 732 | WP_013023830 | conjugal transfer complement resistance protein TraT | - |
| HV026_RS26170 (HV026_26185) | 102497..103186 | + | 690 | WP_165457226 | hypothetical protein | - |
| HV026_RS26175 (HV026_26190) | 103313..105595 | + | 2283 | WP_040215031 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 5582 | GenBank | NZ_CP055594 |
| Plasmid name | pRHBSTW-00001_2 | Incompatibility group | IncFIB |
| Plasmid size | 116760 bp | Coordinate of oriT [Strand] | 76153..76202 [-] |
| Host baterium | Klebsiella pneumoniae strain RHBSTW-00001 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIE9 |