Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104991 |
| Name | oriT_pUSA08-1-SUR15 |
| Organism | Staphylococcus aureus strain USA300-SUR15 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP014419 (20791..20826 [+], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 4..10, 14..20 (GCGAACT..AGTTCGC) |
| Location of nic site | 29..30 |
| Conserved sequence flanking the nic site |
GTGCGCCCTT |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pUSA08-1-SUR15
CACGCGAACTGAAAGTTCGCATAAGTGCGCCCTTAC
CACGCGAACTGAAAGTTCGCATAAGTGCGCCCTTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file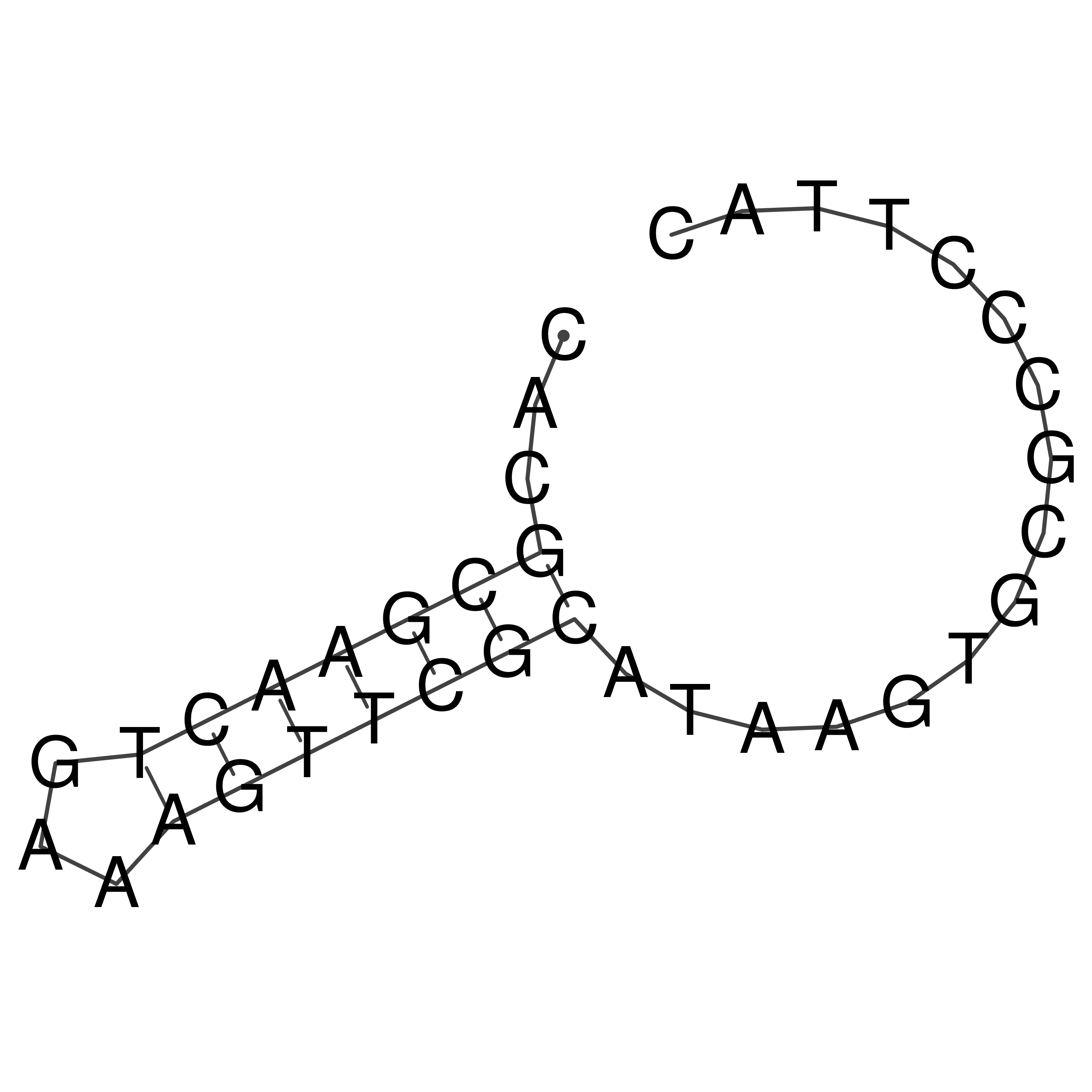
Relaxase
| ID | 3501 | GenBank | WP_077444064 |
| Name | MobA_MobL_AYM28_RS15785_pUSA08-1-SUR15 |
UniProt ID | _ |
| Length | 665 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 665 a.a. Molecular weight: 79052.11 Da Isoelectric Point: 5.8224
>WP_077444064.1 MobA/MobL family protein [Staphylococcus aureus]
MAMYHFQNKFVSKAKGQSATAKSAYNSASKIKDYKENEFKDYSNKQCDYSEILLPYNADEKFKDREYLWN
KVNEVENRKNSQVAREIIIGLPNEFDPKSNVELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWETVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSTISKHNEEIKKRNNSKIDKMFKDVEDIKSHKLNSFSYMQNTDSKILKDMAKDLKLFI
TPINIYKENERLYDLKQKTALISDDEDRLSKIEDIEDRQQKLESINNVFEKQAQIFFDKNYPDQTKDYDK
NEKIFITRTILNDRDDLPAHNELDDIIKDKREKEAQISLNTVLGNRDISLQSIEDESNFFTGKLSHILEE
NSLSFDDVLDNKHEGLDDEMKIDYYTNKLEVLRNAENTLEEYYDIQIKDLFTDNDDYRSFNEVTNIKEKQ
QLIDFKEYHGTENTLEMLETGQFIPKYNDEDRKYITESIKLLQEKEFKPNKSQHDTYVIGAIQKKLLKDF
DFDYSNNNDLKHLYQETNEVKDELSKGNIEEYYNGNDILIDKENFNNYQKKSQASNLINASVDSFIFNFN
EIFRERMPKYINKQYKGKNHSRKRHELKNKRGIHL
MAMYHFQNKFVSKAKGQSATAKSAYNSASKIKDYKENEFKDYSNKQCDYSEILLPYNADEKFKDREYLWN
KVNEVENRKNSQVAREIIIGLPNEFDPKSNVELAKEFAESLSNEGMIVDLNIHKINEENPHAHLLCTLRG
LDKNNEFEPKRKGNDYIRDWNTKEKHNEWRKRWETVQNKHLEKNGFSVRVSADSYKNQNIDLEPTKKEGW
KARKFEDETGKKSTISKHNEEIKKRNNSKIDKMFKDVEDIKSHKLNSFSYMQNTDSKILKDMAKDLKLFI
TPINIYKENERLYDLKQKTALISDDEDRLSKIEDIEDRQQKLESINNVFEKQAQIFFDKNYPDQTKDYDK
NEKIFITRTILNDRDDLPAHNELDDIIKDKREKEAQISLNTVLGNRDISLQSIEDESNFFTGKLSHILEE
NSLSFDDVLDNKHEGLDDEMKIDYYTNKLEVLRNAENTLEEYYDIQIKDLFTDNDDYRSFNEVTNIKEKQ
QLIDFKEYHGTENTLEMLETGQFIPKYNDEDRKYITESIKLLQEKEFKPNKSQHDTYVIGAIQKKLLKDF
DFDYSNNNDLKHLYQETNEVKDELSKGNIEEYYNGNDILIDKENFNNYQKKSQASNLINASVDSFIFNFN
EIFRERMPKYINKQYKGKNHSRKRHELKNKRGIHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3451 | GenBank | WP_077444030 |
| Name | t4cp2_AYM28_RS15680_pUSA08-1-SUR15 |
UniProt ID | _ |
| Length | 543 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 543 a.a. Molecular weight: 62085.11 Da Isoelectric Point: 8.3315
>WP_077444030.1 VirD4-like conjugal transfer protein, CD1115 family [Staphylococcus aureus]
MTTILGELDAKSVKQKKLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGELYE
KTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDASIVAELIIKSAGDSKINRDVWFKSSVGLLNS
LILYAKYEFPPEKRNMENVIKFVQNNKPKKESDDGSAELDKRFSELPKDHPARESYEFGFDVSEDRTRSS
IVLTFVSDLRNYVDREVRSYTSDSDFDLKDVGKQKTIVYVMLPVLGNSNTALSSLFFSQMFQQLYRLGDD
NGAKLPYPVDFLLDEWPNIGAIPDYEETLATCRKYGIGISTIVQSISQVIDKYNKDKANAIIGNHAVILC
LGNVNNDTAKYIKEELGNTTVEFETSSEGSSSGKDSRSKSSNKNVSFTGRALLNEDEISNMELDKAVLIT
KNRNPKIIKKCAYFKMFPDLTEKYLAPQNEYKRKKNASMIERHNKLQKEWDIKQEKLKQKKIEEYKEKER
HQKELEEEQNQVNEVSQNKSEITKSEDEETKTVSDTKKSFYDKLKQKQNANKK
MTTILGELDAKSVKQKKLVIQDFDTKCPNRNIFVVGGPGSFKSAGYVIPNVIVKNQQSIVVTDPSGELYE
KTANIKRMQGYDVRVVNFKNFLASDRQNFFDYIDKDSDASIVAELIIKSAGDSKINRDVWFKSSVGLLNS
LILYAKYEFPPEKRNMENVIKFVQNNKPKKESDDGSAELDKRFSELPKDHPARESYEFGFDVSEDRTRSS
IVLTFVSDLRNYVDREVRSYTSDSDFDLKDVGKQKTIVYVMLPVLGNSNTALSSLFFSQMFQQLYRLGDD
NGAKLPYPVDFLLDEWPNIGAIPDYEETLATCRKYGIGISTIVQSISQVIDKYNKDKANAIIGNHAVILC
LGNVNNDTAKYIKEELGNTTVEFETSSEGSSSGKDSRSKSSNKNVSFTGRALLNEDEISNMELDKAVLIT
KNRNPKIIKKCAYFKMFPDLTEKYLAPQNEYKRKKNASMIERHNKLQKEWDIKQEKLKQKKIEEYKEKER
HQKELEEEQNQVNEVSQNKSEITKSEDEETKTVSDTKKSFYDKLKQKQNANKK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 5429 | GenBank | NZ_CP014419 |
| Plasmid name | pUSA08-1-SUR15 | Incompatibility group | - |
| Plasmid size | 32098 bp | Coordinate of oriT [Strand] | 20791..20826 [+] |
| Host baterium | Staphylococcus aureus strain USA300-SUR15 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA15, AcrIIA21 |