Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104957 |
| Name | oriT_pSA20083530.1 |
| Organism | Salmonella enterica strain SA20083530 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP030204 (71842..71923 [-], 82 nt) |
| oriT length | 82 nt |
| IRs (inverted repeats) | 44..49, 52..57 (CCGCGC..GCGCGG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 82 nt
>oriT_pSA20083530.1
CGGGGTGTCGGGGCGAAGCCCTGACCAGGTGGTAAATGTATGGCCGCGCGTGCGCGGTTATACATTTACACATCCTGTCCCG
CGGGGTGTCGGGGCGAAGCCCTGACCAGGTGGTAAATGTATGGCCGCGCGTGCGCGGTTATACATTTACACATCCTGTCCCG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file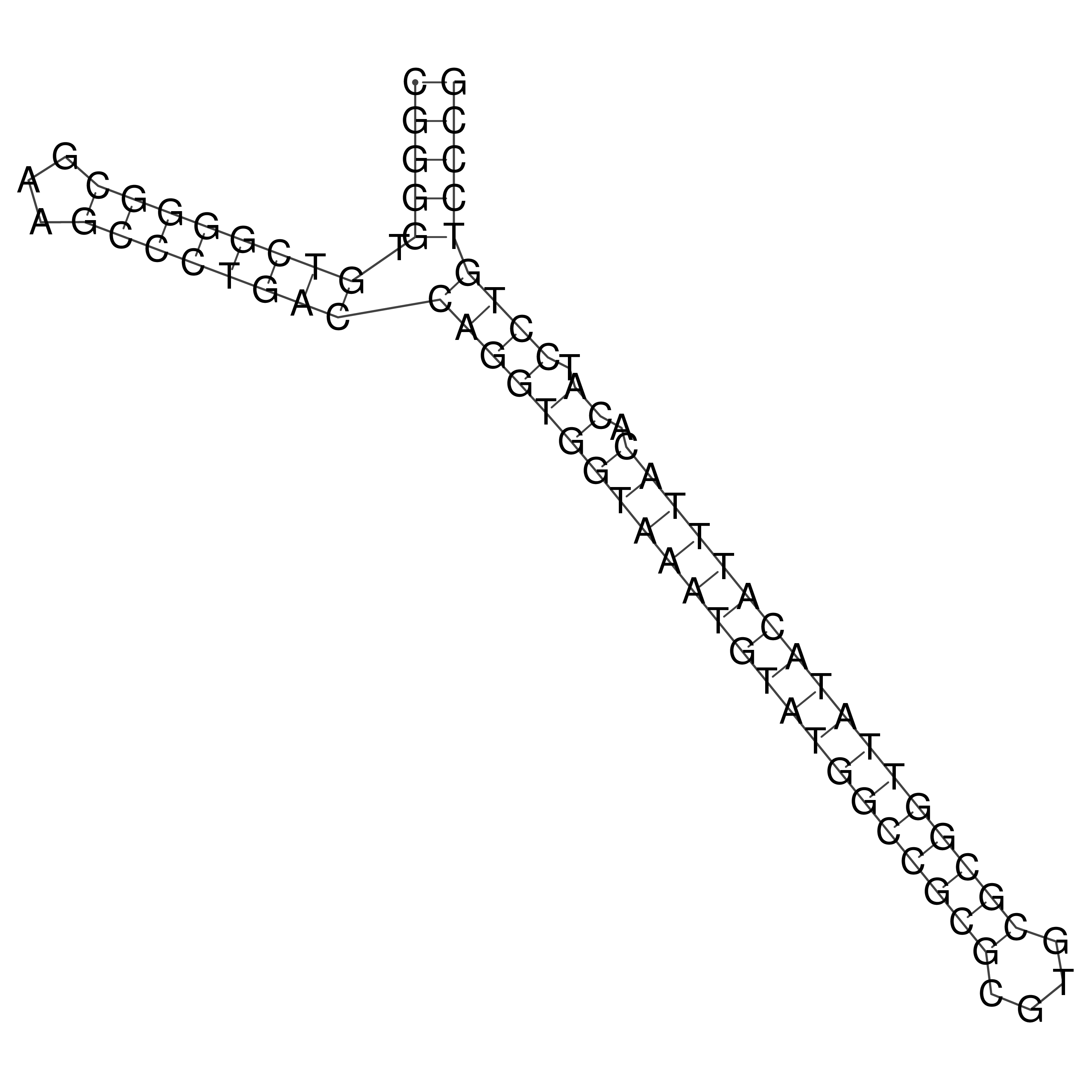
Relaxase
| ID | 3483 | GenBank | WP_114047640 |
| Name | nikB_CHD70_RS27305_pSA20083530.1 |
UniProt ID | _ |
| Length | 899 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 899 a.a. Molecular weight: 104105.46 Da Isoelectric Point: 7.2080
>WP_114047640.1 IncI1-type relaxase NikB [Salmonella enterica]
MNAIIPKKRRDGKSSFEDLVSYVSVRDDVEDEELHALSSVQSELSHRSRFSRLVDYATRLRDESFVSLVD
VMKDGCEWVNFYGVTCFHNCTSLETAAEEMEYTARQARYAKDDTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKALGLPEHQYVSAVHTDTDNLHVHVAVNRVHPDTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
IHAPGNRIVRKTAVERDRQNAWKRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQQNGS
FMVTDGWDRNREGVQLDSFGPSWSGEKLRKKMGEYTPVPNDIFSQVVTPGRYKPEAVAADTRPEKIAETE
SLMQYACRHTGERLPEMAREGQLESCQAIHRTLAEAGLWMRIQHGHLVICDGYDHNQTPVRADSVWSLLT
LENVKQLSGGWQPVPTDIFRQVTPTERFSGRRLEICPASDKEWHRMRTGTGPQGAIRRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLARAEP
QAGPFVSAPADLFDRVKPESRYNPELAVSDKYGISSKRDPMLRRQRREARAEARADLRARYLAWRTQWCK
PDLRYGERCRDIHQACRLRKSHIRAQYDDPVLRKLHYHIAEVQRMQALIRLKEEVRDERQKLIADGKWYP
PSYRQWVETQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTADRCVILCEPGGTPVYENRGDLEARLQKNGS
VRFRDRRTDQFVCTDYGDRVVFHNHHDRNELADKLDLIAPVLFERDPRIGFEPEGNDRQFNQVFAEMVAW
HNVTERTGHGDYTISRPDVDHHREGCERYYRDYIDANSSNETSLPSPEQEKRWEPPSPG
MNAIIPKKRRDGKSSFEDLVSYVSVRDDVEDEELHALSSVQSELSHRSRFSRLVDYATRLRDESFVSLVD
VMKDGCEWVNFYGVTCFHNCTSLETAAEEMEYTARQARYAKDDTDPVFHYILSWQAHESPRPEQIYDSVR
HTLKALGLPEHQYVSAVHTDTDNLHVHVAVNRVHPDTGYLNCLSWSQEKLSRACRELELKHGFAPDNGCW
IHAPGNRIVRKTAVERDRQNAWKRGKKQTFREYVAQTAVAGLRSEPVHDWLSLHRRLAEDGLYLSQQNGS
FMVTDGWDRNREGVQLDSFGPSWSGEKLRKKMGEYTPVPNDIFSQVVTPGRYKPEAVAADTRPEKIAETE
SLMQYACRHTGERLPEMAREGQLESCQAIHRTLAEAGLWMRIQHGHLVICDGYDHNQTPVRADSVWSLLT
LENVKQLSGGWQPVPTDIFRQVTPTERFSGRRLEICPASDKEWHRMRTGTGPQGAIRRELFSDKESLWGY
SISHCSPQIEEMITQGEFTWQRCHELFAQQGLMLQKQHHGLVVVDAFNHEQTPVKASSIHPDLTLARAEP
QAGPFVSAPADLFDRVKPESRYNPELAVSDKYGISSKRDPMLRRQRREARAEARADLRARYLAWRTQWCK
PDLRYGERCRDIHQACRLRKSHIRAQYDDPVLRKLHYHIAEVQRMQALIRLKEEVRDERQKLIADGKWYP
PSYRQWVETQAAQGDRAAVSQLRGWDYRDRRKDKSRTTTADRCVILCEPGGTPVYENRGDLEARLQKNGS
VRFRDRRTDQFVCTDYGDRVVFHNHHDRNELADKLDLIAPVLFERDPRIGFEPEGNDRQFNQVFAEMVAW
HNVTERTGHGDYTISRPDVDHHREGCERYYRDYIDANSSNETSLPSPEQEKRWEPPSPG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1551 | GenBank | WP_114047639 |
| Name | WP_114047639_pSA20083530.1 |
UniProt ID | _ |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12699.62 Da Isoelectric Point: 10.7418
>WP_114047639.1 plasmid mobilization protein MobA [Salmonella enterica]
MSDKRERSGSERRQKTLLRAVRFSPDEDEIIRKKAEDAGLTVSAYIRSAALNTRVNSRIDDKFLKELMRL
GRMQKHLFVEGKRTGDKEYASVLVAITELSNTVRKKLMEN
MSDKRERSGSERRQKTLLRAVRFSPDEDEIIRKKAEDAGLTVSAYIRSAALNTRVNSRIDDKFLKELMRL
GRMQKHLFVEGKRTGDKEYASVLVAITELSNTVRKKLMEN
Protein domains
No domain identified.
Protein structure
No available structure.
T4CP
| ID | 3430 | GenBank | WP_114047645 |
| Name | trbC_CHD70_RS27330_pSA20083530.1 |
UniProt ID | _ |
| Length | 763 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 763 a.a. Molecular weight: 86727.96 Da Isoelectric Point: 7.4501
>WP_114047645.1 F-type conjugative transfer protein TrbC [Salmonella enterica]
MSEHRVNPELIHRAAWGNPLWNALQTLNIYGLCLAGSLVVSFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLKYADPSQDRMIKRSLFSFWPTLFQYEVIKESPANGIFYVGYQRVRDIGRELWLNIDDLTRHIM
FFATTGGGKTETIFAWAINPLCWGRGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTETFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFVTQQSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNSLQRQDGIFGESWIDSPQISILMESKINVQELIELH
AGEFFSIFRGDTVPSASFLIPDGEKSCSSDPVVINRYISVDAPHLDCLRRLVPRTAQRRIPSPENVSAIT
GVLTAKPSRKRRKIRTELHTIVDTFQQRIAGRQAAMAMLEEYDTDINARERALWETAVNTLKTTTREERR
IRYITLNRPDTPATEEENQISVRTERAGINLLALPKDNKHPAGRPVSGLHHKKNRRPDWDGMY
MSEHRVNPELIHRAAWGNPLWNALQTLNIYGLCLAGSLVVSFIWPLALPACLLFTLITMLVFSLQRWRCP
LRMPMTLKYADPSQDRMIKRSLFSFWPTLFQYEVIKESPANGIFYVGYQRVRDIGRELWLNIDDLTRHIM
FFATTGGGKTETIFAWAINPLCWGRGFTLVDGKAQNDTARTIWYLARRFGREDDVEVINFMNGGKSRSEI
ILSGEKTRPQSNTWNPFCYSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVREGKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFSTFTETFGDIFAEDSGDIDIRDSIHSDRILMVMIPALDTSAHTTSALGRMFVTQQSMILARDLGYRLE
GTDSDALEVKKYKGRFPYLCFLDEVGAYYTDRIAVEATQVRSLDFALILMAQDQERIEGQTTATNTATLM
QNTGTKFAGRIVSEGSTARTLKSAAGEEARARMNSLQRQDGIFGESWIDSPQISILMESKINVQELIELH
AGEFFSIFRGDTVPSASFLIPDGEKSCSSDPVVINRYISVDAPHLDCLRRLVPRTAQRRIPSPENVSAIT
GVLTAKPSRKRRKIRTELHTIVDTFQQRIAGRQAAMAMLEEYDTDINARERALWETAVNTLKTTTREERR
IRYITLNRPDTPATEEENQISVRTERAGINLLALPKDNKHPAGRPVSGLHHKKNRRPDWDGMY
Protein domains
No domain identified.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 81091..119367
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CHD70_RS27315 (CHD70_27325) | 76385..76762 | - | 378 | WP_114047642 | RidA family protein | - |
| CHD70_RS27320 (CHD70_27330) | 76785..77744 | - | 960 | WP_114047643 | sugar phosphate isomerase/epimerase family protein | - |
| CHD70_RS27325 (CHD70_27335) | 77930..78613 | + | 684 | WP_114047644 | PAS domain-containing protein | - |
| CHD70_RS27330 (CHD70_27340) | 78807..81098 | - | 2292 | WP_114047645 | F-type conjugative transfer protein TrbC | - |
| CHD70_RS27335 (CHD70_27345) | 81091..82161 | - | 1071 | WP_114047646 | DsbC family protein | trbB |
| CHD70_RS27340 (CHD70_27350) | 82180..83388 | - | 1209 | WP_114047647 | IncI1-type conjugal transfer protein TrbA | trbA |
| CHD70_RS27345 (CHD70_27355) | 83536..83880 | + | 345 | WP_080102217 | toxin | - |
| CHD70_RS27350 (CHD70_27360) | 83873..84187 | + | 315 | WP_114047648 | type II toxin-antitoxin system MqsA family antitoxin | - |
| CHD70_RS27355 (CHD70_27365) | 84301..85610 | - | 1310 | Protein_100 | IS3 family transposase | - |
| CHD70_RS27360 (CHD70_27370) | 85644..85778 | - | 135 | Protein_101 | Hha/YmoA family nucleoid-associated regulatory protein | - |
| CHD70_RS27365 (CHD70_27375) | 86008..86655 | - | 648 | WP_205318806 | plasmid IncI1-type surface exclusion protein ExcA | - |
| CHD70_RS27370 (CHD70_27380) | 86735..87040 | - | 306 | Protein_103 | hypothetical protein | - |
| CHD70_RS27375 (CHD70_27385) | 87060..88091 | - | 1032 | WP_114046824 | IS630 family transposase | - |
| CHD70_RS27380 (CHD70_27390) | 88192..90060 | - | 1869 | Protein_105 | DotA/TraY family protein | - |
| CHD70_RS27385 (CHD70_27395) | 90159..90743 | - | 585 | WP_114047651 | conjugal transfer protein TraX | - |
| CHD70_RS27395 (CHD70_27405) | 90984..92186 | - | 1203 | WP_114047653 | conjugal transfer protein TraW | traW |
| CHD70_RS27400 (CHD70_27410) | 92144..92773 | - | 630 | WP_114047654 | conjugal transfer protein TraV | - |
| CHD70_RS27405 (CHD70_27415) | 92773..95817 | - | 3045 | WP_114047655 | ATP-binding protein | traU |
| CHD70_RS27410 (CHD70_27420) | 95915..96715 | - | 801 | WP_114047656 | conjugal transfer protein TraT | traT |
| CHD70_RS27415 (CHD70_27425) | 96699..96887 | - | 189 | WP_000556965 | hypothetical protein | - |
| CHD70_RS27420 (CHD70_27430) | 96956..97360 | - | 405 | WP_080102228 | DUF6750 family protein | traR |
| CHD70_RS27425 (CHD70_27435) | 97403..97930 | - | 528 | WP_080102229 | conjugal transfer protein TraQ | traQ |
| CHD70_RS27430 (CHD70_27440) | 97930..98643 | - | 714 | WP_114047657 | conjugal transfer protein TraP | traP |
| CHD70_RS27435 (CHD70_27445) | 98640..99941 | - | 1302 | WP_114047658 | conjugal transfer protein TraO | traO |
| CHD70_RS27440 (CHD70_27450) | 99944..100927 | - | 984 | WP_114047659 | DotH/IcmK family type IV secretion protein | traN |
| CHD70_RS27445 (CHD70_27455) | 100943..101566 | - | 624 | WP_080102285 | DotI/IcmL family type IV secretion protein | traM |
| CHD70_RS27450 (CHD70_27460) | 101641..101991 | - | 351 | WP_080102233 | conjugal transfer protein | traL |
| CHD70_RS27455 (CHD70_27465) | 102009..105560 | - | 3552 | WP_114047660 | LPD7 domain-containing protein | - |
| CHD70_RS27460 (CHD70_27470) | 105840..106397 | - | 558 | WP_114047661 | phospholipase D family protein | - |
| CHD70_RS29205 (CHD70_27475) | 106412..106702 | - | 291 | WP_001838506 | hypothetical protein | traK |
| CHD70_RS27470 (CHD70_27480) | 106699..107847 | - | 1149 | WP_114047662 | plasmid transfer ATPase TraJ | virB11 |
| CHD70_RS27475 (CHD70_27485) | 107991..108824 | - | 834 | WP_080102237 | type IV secretory system conjugative DNA transfer family protein | traI |
| CHD70_RS27480 (CHD70_27490) | 108821..109276 | - | 456 | WP_114047663 | DotD/TraH family lipoprotein | - |
| CHD70_RS27485 (CHD70_27495) | 109797..110999 | - | 1203 | WP_080102240 | conjugal transfer protein TraF | - |
| CHD70_RS27490 (CHD70_27500) | 111086..111910 | - | 825 | WP_114047664 | conjugal transfer protein TraE | traE |
| CHD70_RS27495 (CHD70_27505) | 112061..113215 | - | 1155 | WP_114047665 | site-specific integrase | - |
| CHD70_RS29210 (CHD70_27510) | 113267..113524 | + | 258 | Protein_128 | Shufflon protein B | - |
| CHD70_RS27505 (CHD70_27515) | 113521..114870 | - | 1350 | WP_240198389 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| CHD70_RS27510 (CHD70_27520) | 114858..115514 | - | 657 | WP_114047666 | A24 family peptidase | - |
| CHD70_RS27515 (CHD70_27525) | 115499..116059 | - | 561 | WP_114047667 | lytic transglycosylase domain-containing protein | virB1 |
| CHD70_RS27520 (CHD70_27530) | 116069..116686 | - | 618 | WP_114047668 | type 4 pilus major pilin | - |
| CHD70_RS27525 (CHD70_27535) | 116704..117801 | - | 1098 | WP_114047669 | type II secretion system F family protein | - |
| CHD70_RS27530 (CHD70_27540) | 117814..119367 | - | 1554 | WP_080102248 | ATPase, T2SS/T4P/T4SS family | virB11 |
| CHD70_RS27535 (CHD70_27545) | 119378..119830 | - | 453 | WP_114047670 | type IV pilus biogenesis protein PilP | - |
| CHD70_RS27540 (CHD70_27550) | 119817..121118 | - | 1302 | WP_114047671 | type 4b pilus protein PilO2 | - |
| CHD70_RS27545 (CHD70_27555) | 121111..122793 | - | 1683 | WP_114047672 | PilN family type IVB pilus formation outer membrane protein | - |
| CHD70_RS27550 (CHD70_27560) | 122807..123244 | - | 438 | WP_080102252 | type IV pilus biogenesis protein PilM | - |
| CHD70_RS27555 (CHD70_27565) | 123244..124311 | - | 1068 | WP_114047673 | TcpQ domain-containing protein | - |
Host bacterium
| ID | 5395 | GenBank | NZ_CP030204 |
| Plasmid name | pSA20083530.1 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 138648 bp | Coordinate of oriT [Strand] | 71842..71923 [-] |
| Host baterium | Salmonella enterica strain SA20083530 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |