Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104881 |
| Name | oriT_pRGM101a |
| Organism | Rhizobium gallicum strain M101 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP092345 (225639..225672 [-], 34 nt) |
| oriT length | 34 nt |
| IRs (inverted repeats) | 17..22, 27..32 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 34 nt
>oriT_pRGM101a
CAAGGGCGCAATTATACGTCGCTGACGCGACGCC
CAAGGGCGCAATTATACGTCGCTGACGCGACGCC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file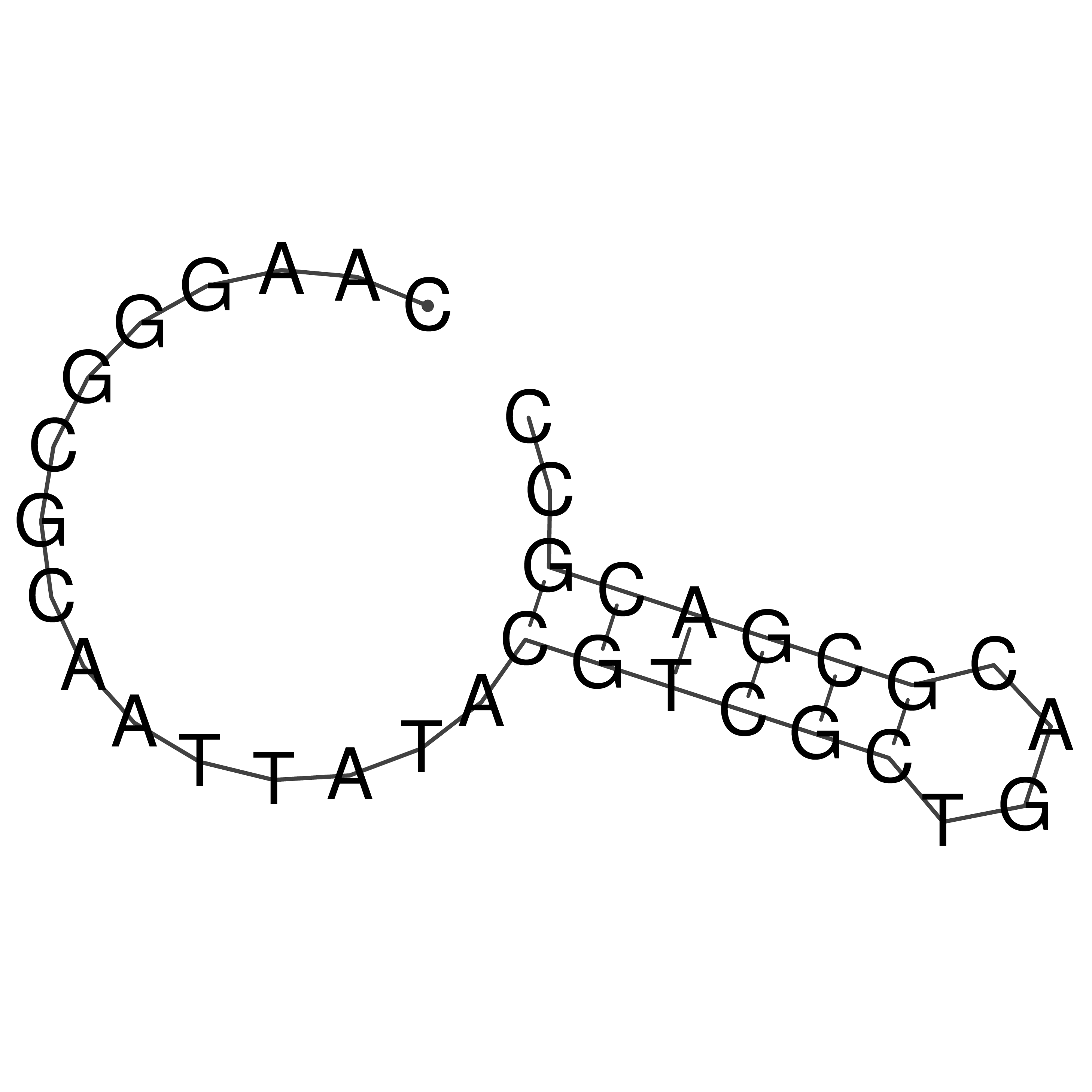
Relaxase
| ID | 3453 | GenBank | WP_239600232 |
| Name | traA_L2W42_RS22100_pRGM101a |
UniProt ID | _ |
| Length | 1103 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1103 a.a. Molecular weight: 122478.83 Da Isoelectric Point: 10.0051
>WP_239600232.1 Ti-type conjugative transfer relaxase TraA [Rhizobium gallicum]
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTCKQGLAHEEFVIPADAPDWLRTIIADR
SVPGASEAFWNKVEAFEKRSDAQLAKDVTIALPVELTYEQNIALVREFVERHIIATGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGAKKVALLGPDGNPIRNDSGKIVYELWAGSADDFNALRDGWFACQNRHLALAG
LDISIDGRSFEKQGIKLTPTIHLGVRTKAIERKSEAATKPLPLERIELQEDRRAENARRVQRHPEIVLDL
ITREKSVFDERDVAKVLHRYIDDADLFQSLLARTLQSPETLQLESERIEFATGVRVSAKHTTREVIQLEA
EMANRAIWLSHRSSHGVREAVLEATFSRHSRLSEEQKIAIVHVAGGERIAAVIGRAGVGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGITSRTLASWELRWSEGRNQLDDKTVFVLDEAGMVSSRQMALFIE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRKQWMRDASLDLARGKVGKAVDAYR
AYGRFKGSELKAQAVENLIADWNGDYDPVKTTLILAHLRRDVRMLNEMARAKLIERGILAEGFVFKTEDG
NRKFAAGDQIVFLKNEGSLGVKNGMLAKVVEALPGRIVAEIGEGEHRRQVRVEQRFYNKVDHGYATTIHK
SQGATVDRVKILASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSDGLIPILSRRNAKETTLDYEKGPFY
RQALRFAEARGLRLVNVARMLVRDRHEWSIRQKQKLADLAARLAAIGGKLSLSTSAKNSPNQKIIKEVKP
MVSGIATFAKSLEQAVEDKVATDPGLEKQWEEVSSRFHLVYAQPETAFKAINVDAMLKDKATAQSTISRI
GTQPESFGALKGKTGILASRADKQGREKALTNAPALARNIERYLRQRAEAERKHEVEERAVRLKVSVNIP
ALSPAAKQALERVRDAIDRNDLPVGLEYALAAKMVKAELEGFARAVSERFGERTFLPLAAKDTSGQAFQT
VTAGMSAGQKAEVQSAWNSMRTVQQFAAHERTTEALKQAETLRQTKAQGLSLK
MAVPHFSVSIVARGSGRSAVLSAAYRHCAKMDYEREARTIDYTCKQGLAHEEFVIPADAPDWLRTIIADR
SVPGASEAFWNKVEAFEKRSDAQLAKDVTIALPVELTYEQNIALVREFVERHIIATGMVADWVYHDAPGN
PHVHLMTTLRPLTDDGFGAKKVALLGPDGNPIRNDSGKIVYELWAGSADDFNALRDGWFACQNRHLALAG
LDISIDGRSFEKQGIKLTPTIHLGVRTKAIERKSEAATKPLPLERIELQEDRRAENARRVQRHPEIVLDL
ITREKSVFDERDVAKVLHRYIDDADLFQSLLARTLQSPETLQLESERIEFATGVRVSAKHTTREVIQLEA
EMANRAIWLSHRSSHGVREAVLEATFSRHSRLSEEQKIAIVHVAGGERIAAVIGRAGVGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGITSRTLASWELRWSEGRNQLDDKTVFVLDEAGMVSSRQMALFIE
AATKAGAKLVLVGDPEQLQPIEAGAAFRAIADRIGYAELETIYRQRKQWMRDASLDLARGKVGKAVDAYR
AYGRFKGSELKAQAVENLIADWNGDYDPVKTTLILAHLRRDVRMLNEMARAKLIERGILAEGFVFKTEDG
NRKFAAGDQIVFLKNEGSLGVKNGMLAKVVEALPGRIVAEIGEGEHRRQVRVEQRFYNKVDHGYATTIHK
SQGATVDRVKILASLSLDRHLTYVAMTRHREDLAVYYGRRSFAKSDGLIPILSRRNAKETTLDYEKGPFY
RQALRFAEARGLRLVNVARMLVRDRHEWSIRQKQKLADLAARLAAIGGKLSLSTSAKNSPNQKIIKEVKP
MVSGIATFAKSLEQAVEDKVATDPGLEKQWEEVSSRFHLVYAQPETAFKAINVDAMLKDKATAQSTISRI
GTQPESFGALKGKTGILASRADKQGREKALTNAPALARNIERYLRQRAEAERKHEVEERAVRLKVSVNIP
ALSPAAKQALERVRDAIDRNDLPVGLEYALAAKMVKAELEGFARAVSERFGERTFLPLAAKDTSGQAFQT
VTAGMSAGQKAEVQSAWNSMRTVQQFAAHERTTEALKQAETLRQTKAQGLSLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3398 | GenBank | WP_239600215 |
| Name | t4cp2_L2W42_RS22020_pRGM101a |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91381.92 Da Isoelectric Point: 5.7875
>WP_239600215.1 conjugal transfer protein TrbE [Rhizobium gallicum]
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRVSTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDAESRKKSYADTVLFAFRNAIREIEQYLANTLSIRRMETRETAERGGERIARYDELLQFVRFCITG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQNGTVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNAADAFLGSLPGNWYSNIREPLINTSNLADLIPLNAVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNVHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYESAQVFA
FDKGCSLLPLTLATGGDHYEIGGDNAEEGRALTFCPLSDLGSDADRAWATEWVEMLVGLQGVAITPDHRN
AISRQVGLMASAAGRSISDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLGTFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLNGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESSPTKICLPNGAARESGTREFYERIGFNERQIEIVSSAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLQTKGVRDAASLLNFE
MVALKRFRATGPSFADLVPYAGLVDNGVLLLKDGSLMAGWYFAGPDSESATDLERNELSRQINAVLSRLG
SGWMIQVEAIRVSTVDYPSEDRCHFPDPVTRAIDAERRAHFAREQGHFESKHALILTYRPLESKKTALSK
YIYSDAESRKKSYADTVLFAFRNAIREIEQYLANTLSIRRMETRETAERGGERIARYDELLQFVRFCITG
ESHPIRLPDVPMYLDWIATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDLMPLTYRWSSRFIF
LDAEEARQKLERTRKKWQQKVRPFFDQLFQTQNGTVDQDAMTMVAETEDAIAQASSQLVAYGYYTPVVVL
FDHDREALQEKAEAIRRLIQAEGFGARIETLNAADAFLGSLPGNWYSNIREPLINTSNLADLIPLNAVWS
GNPVAPCPFYPPNSPPLMQVASGSTPFRLNVHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYESAQVFA
FDKGCSLLPLTLATGGDHYEIGGDNAEEGRALTFCPLSDLGSDADRAWATEWVEMLVGLQGVAITPDHRN
AISRQVGLMASAAGRSISDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLGTFQTFEIEQLMNMG
ERNLVPVLTYLFRRIEKRLNGSPSLIVLDEAWLMLGHPVFRSKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESSPTKICLPNGAARESGTREFYERIGFNERQIEIVSSAIPKREYYVATPDGRRLFDMSLGP
VALSFVGASGKEDLKRIRALKSEHGHDWPIHWLQTKGVRDAASLLNFE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3399 | GenBank | WP_239600240 |
| Name | traG_L2W42_RS22115_pRGM101a |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 69063.39 Da Isoelectric Point: 9.2211
>WP_239600240.1 Ti-type conjugative transfer system protein TraG [Rhizobium gallicum]
MNRVLLAITPIMLMVGVAILASGMAPSLAGFGKTEAAKLLLGRAGIALPYVLAAAIGVVFLFASVGAIRI
HFAGWGVVAGSASAIAIAALREATRLAAFAEQLPAGRFIFAYLDPATMVGAAAALMAGCFALRVALLGNA
AFARSEPTRIRGKRAMHGEADWMKLNEAEKLLPEAGGIVIGERYRVDKDSTAALSFRADSPETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSDEVAPMVCKRREDAGRRIRILDPR
KPQTGFNALDWIGKFGGTKEEDIAAVASWIMSDSGRASGVREDFFRASGLQLLTAIIADVCLSGHTEKEL
QTLRQVRANLSEPEPKLRQRLQDIYDNSVSPFVKENVAAFVNMTPETFSGVYANAIKETHWLSYPNYAVL
VSGSTFLTEDIASGETDVFINIDLKALETHAGLARVIIGSFVNAIYNRDGVMNGRALFLLDEVARLGHMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDATSKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSFQAKGSSRTRSKQLTARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRQDMRIWVAEN
RFESPST
MNRVLLAITPIMLMVGVAILASGMAPSLAGFGKTEAAKLLLGRAGIALPYVLAAAIGVVFLFASVGAIRI
HFAGWGVVAGSASAIAIAALREATRLAAFAEQLPAGRFIFAYLDPATMVGAAAALMAGCFALRVALLGNA
AFARSEPTRIRGKRAMHGEADWMKLNEAEKLLPEAGGIVIGERYRVDKDSTAALSFRADSPETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGTLIVLDPSDEVAPMVCKRREDAGRRIRILDPR
KPQTGFNALDWIGKFGGTKEEDIAAVASWIMSDSGRASGVREDFFRASGLQLLTAIIADVCLSGHTEKEL
QTLRQVRANLSEPEPKLRQRLQDIYDNSVSPFVKENVAAFVNMTPETFSGVYANAIKETHWLSYPNYAVL
VSGSTFLTEDIASGETDVFINIDLKALETHAGLARVIIGSFVNAIYNRDGVMNGRALFLLDEVARLGHMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDATSKWFESASWISFAAINDPETADYISRRCGMTTVE
IDQVSRSFQAKGSSRTRSKQLTARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRQDMRIWVAEN
RFESPST
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 201643..217562
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| L2W42_RS21960 (L2W42_21960) | 197759..198744 | + | 986 | Protein_201 | plasmid partitioning protein RepB | - |
| L2W42_RS21965 (L2W42_21965) | 200091..200531 | + | 441 | WP_239600196 | hypothetical protein | - |
| L2W42_RS21970 (L2W42_21970) | 200605..201306 | + | 702 | WP_239600198 | IS6 family transposase | - |
| L2W42_RS21975 (L2W42_21975) | 201375..201629 | + | 255 | Protein_204 | nitrilase-related carbon-nitrogen hydrolase | - |
| L2W42_RS21980 (L2W42_21980) | 201643..202290 | + | 648 | WP_239600200 | TraH family protein | virB1 |
| L2W42_RS21985 (L2W42_21985) | 203330..204544 | - | 1215 | WP_239600202 | plasmid replication protein RepC | - |
| L2W42_RS21990 (L2W42_21990) | 204746..205771 | - | 1026 | WP_239600204 | plasmid partitioning protein RepB | - |
| L2W42_RS21995 (L2W42_21995) | 205768..207030 | - | 1263 | WP_239600278 | plasmid partitioning protein RepA | - |
| L2W42_RS22000 (L2W42_22000) | 207373..207996 | + | 624 | WP_239600206 | acyl-homoserine-lactone synthase TraI | - |
| L2W42_RS22005 (L2W42_22005) | 208007..208984 | + | 978 | WP_239600208 | P-type conjugative transfer ATPase TrbB | virB11 |
| L2W42_RS22010 (L2W42_22010) | 208974..209357 | + | 384 | WP_239600211 | TrbC/VirB2 family protein | virB2 |
| L2W42_RS22015 (L2W42_22015) | 209350..209649 | + | 300 | WP_239600213 | conjugal transfer protein TrbD | virB3 |
| L2W42_RS22020 (L2W42_22020) | 209660..212116 | + | 2457 | WP_239600215 | conjugal transfer protein TrbE | virb4 |
| L2W42_RS22025 (L2W42_22025) | 212088..212891 | + | 804 | WP_239600217 | P-type conjugative transfer protein TrbJ | virB5 |
| L2W42_RS22030 (L2W42_22030) | 212888..213088 | + | 201 | WP_239600219 | entry exclusion protein TrbK | - |
| L2W42_RS22035 (L2W42_22035) | 213082..214263 | + | 1182 | WP_239600220 | P-type conjugative transfer protein TrbL | virB6 |
| L2W42_RS22040 (L2W42_22040) | 214284..214946 | + | 663 | WP_239600221 | conjugal transfer protein TrbF | virB8 |
| L2W42_RS22045 (L2W42_22045) | 214963..215772 | + | 810 | WP_239600222 | P-type conjugative transfer protein TrbG | virB9 |
| L2W42_RS22050 (L2W42_22050) | 215776..216213 | + | 438 | WP_239600224 | conjugal transfer protein TrbH | - |
| L2W42_RS22055 (L2W42_22055) | 216225..217562 | + | 1338 | WP_239600226 | IncP-type conjugal transfer protein TrbI | virB10 |
| L2W42_RS22060 (L2W42_22060) | 217859..218563 | + | 705 | WP_239600280 | autoinducer binding domain-containing protein | - |
| L2W42_RS22065 (L2W42_22065) | 218567..218890 | - | 324 | WP_239600227 | transcriptional repressor TraM | - |
| L2W42_RS22070 (L2W42_22070) | 219107..219319 | + | 213 | WP_239600229 | helix-turn-helix transcriptional regulator | - |
| L2W42_RS22075 (L2W42_22075) | 219444..220072 | + | 629 | Protein_224 | DUF433 domain-containing protein | - |
| L2W42_RS36075 | 220069..220356 | + | 288 | Protein_225 | DUF5615 family PIN-like protein | - |
| L2W42_RS22085 (L2W42_22085) | 220368..220520 | - | 153 | Protein_226 | conjugal transfer protein TraH | - |
| L2W42_RS22090 (L2W42_22090) | 220537..221700 | - | 1164 | WP_239600230 | conjugal transfer protein TraB | - |
| L2W42_RS22095 (L2W42_22095) | 221690..222193 | - | 504 | WP_239600231 | conjugative transfer signal peptidase TraF | - |
Host bacterium
| ID | 5319 | GenBank | NZ_CP092345 |
| Plasmid name | pRGM101a | Incompatibility group | - |
| Plasmid size | 513852 bp | Coordinate of oriT [Strand] | 225639..225672 [-] |
| Host baterium | Rhizobium gallicum strain M101 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | linJ, dxnH, LinG |
| Symbiosis gene | fixS, fixQ, fixO, fixN, nodM, nodD, mLTONO_5203, nifX, nifN, nifK, nifD, nifH, nifS, nifW, fixA, fixB, fixC, fixX, nifA, nifZ, nifT, nodH, nodJ, nodI, nodU, nodS, nodC, nodB, nodA |
| Anti-CRISPR | AcrIF3, AcrIIC1 |