Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104877 |
| Name | oriT_pRSpC104g |
| Organism | Rhizobium sp. C104 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP092347 (176070..176111 [+], 42 nt) |
| oriT length | 42 nt |
| IRs (inverted repeats) | 22..28, 33..39 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 42 nt
>oriT_pRSpC104g
GGATCCGAAGGGCGCAATTATACGTCGCTGGTGCGACGTCCT
GGATCCGAAGGGCGCAATTATACGTCGCTGGTGCGACGTCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file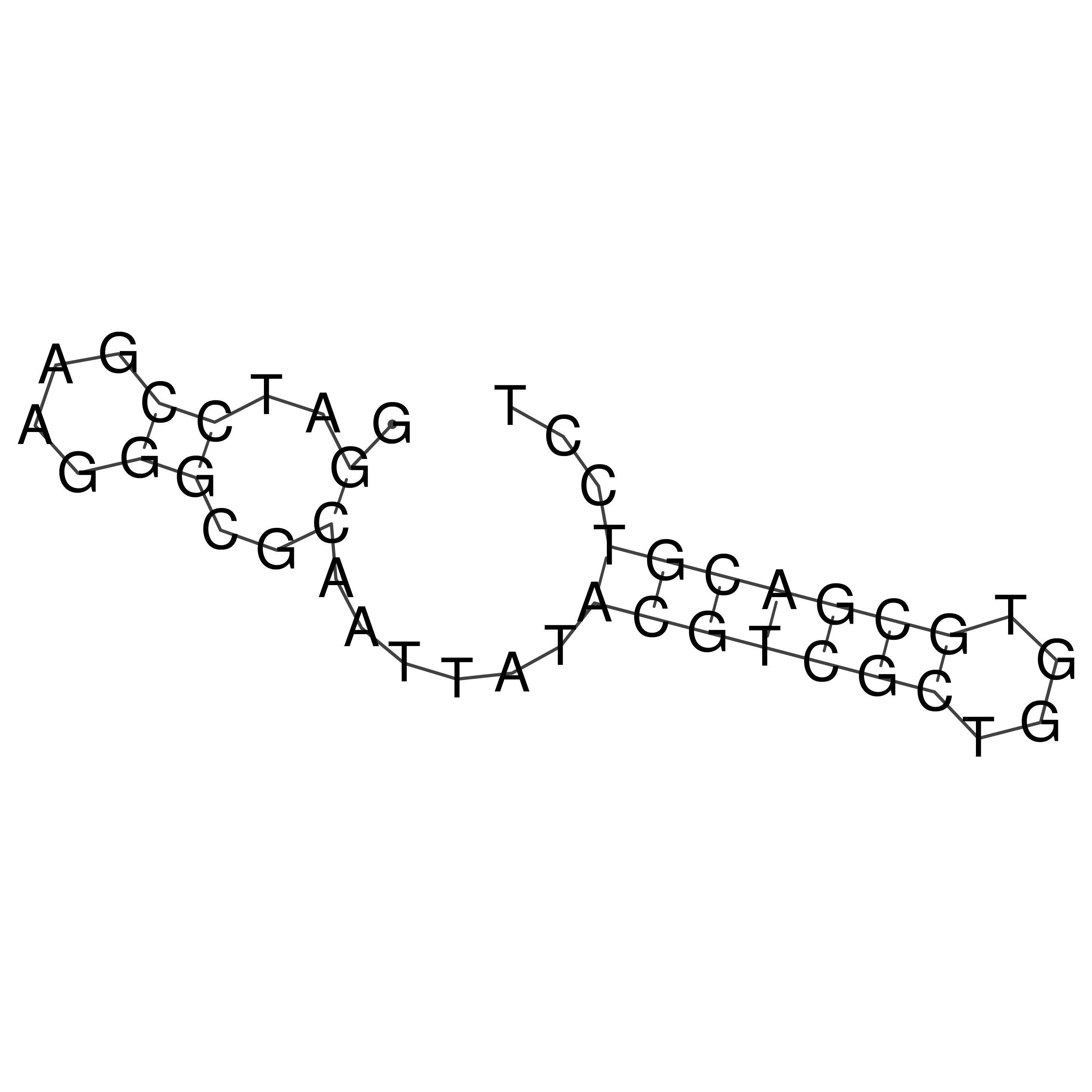
T4CP
| ID | 3393 | GenBank | WP_096772214 |
| Name | t4cp2_MF410_RS00055_pRSpC104g |
UniProt ID | _ |
| Length | 567 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 567 a.a. Molecular weight: 62800.99 Da Isoelectric Point: 7.1079
>WP_096772214.1 MULTISPECIES: ATP-binding protein [Rhizobium]
MIKHSLVDPDRYVGTISVVTASTVQVNLPYATARPERRALARGAVGDFVFVDCELVKLLGRLVEVKLPDA
ERLTVEPTLGTQPDPHPVGRVQLLASVDQRQSKLIRGIRHHPRVGDAVYLADPSMFGELMANSLVASGTS
ALDIGTLDAGSGVHLRLPPEKLFGRHCGVFGATGGGKSWTLATILQQVKAAGGKCILFDPTGEFAEIPAI
SEHYAFDHEEAGSQQVHFPYQFMTEDDLFALFRPSGQSQGPKLREAIKSLKLVKKSEGQLEGVTIYEKKL
LLKKQKDRRPFLKALEVHSDVIHSPLCHFDIENLPDQVQQECVQPTDWDNPTHWGKVNEQAVGYCESLIA
RMRTLIHTPELECLFGNRGQSLVKVLEAFFASERQDILRISFKNVRFEHNTREILLNVIGRFLLARARQN
TFRDKPMITLLDEAHQFLGRTIGDEYASVKLEAFGLIAKEGRKYGLTTILATQRPRDIPADVLSQLGTLI
VHRLTNEDDRNAVEKACGDLDRNAALFIPTLAPGEAIIVGPDLPAPVPVLIHEPPSPPDSKGPKYTSYWA
SRQPRAD
MIKHSLVDPDRYVGTISVVTASTVQVNLPYATARPERRALARGAVGDFVFVDCELVKLLGRLVEVKLPDA
ERLTVEPTLGTQPDPHPVGRVQLLASVDQRQSKLIRGIRHHPRVGDAVYLADPSMFGELMANSLVASGTS
ALDIGTLDAGSGVHLRLPPEKLFGRHCGVFGATGGGKSWTLATILQQVKAAGGKCILFDPTGEFAEIPAI
SEHYAFDHEEAGSQQVHFPYQFMTEDDLFALFRPSGQSQGPKLREAIKSLKLVKKSEGQLEGVTIYEKKL
LLKKQKDRRPFLKALEVHSDVIHSPLCHFDIENLPDQVQQECVQPTDWDNPTHWGKVNEQAVGYCESLIA
RMRTLIHTPELECLFGNRGQSLVKVLEAFFASERQDILRISFKNVRFEHNTREILLNVIGRFLLARARQN
TFRDKPMITLLDEAHQFLGRTIGDEYASVKLEAFGLIAKEGRKYGLTTILATQRPRDIPADVLSQLGTLI
VHRLTNEDDRNAVEKACGDLDRNAALFIPTLAPGEAIIVGPDLPAPVPVLIHEPPSPPDSKGPKYTSYWA
SRQPRAD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3394 | GenBank | WP_096771503 |
| Name | traG_MF410_RS00795_pRSpC104g |
UniProt ID | _ |
| Length | 658 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 658 a.a. Molecular weight: 70846.75 Da Isoelectric Point: 9.1951
>WP_096771503.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium]
MSRIMLFIAPVALMILVAAGLTGIEHWLSGLGKTEAARQTLGRAGIALPYVSAGLVGIVFLFASAGAIRI
KTAGWGVVAGGAATILIAAIRETMRLSALTDKVPAGMSIFSYVDPATLIGAAAAAIAACFALRVALAGNA
AFASAEPKRVRGKRALHGEAEWMKLPEAANLFSDTGGIVIGERYRVDKDSTAARSFRADEPETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSAHRTSANRDLFVLDPK
NPATGFNALDWIGKFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTAMIADVCLSGHTEKEN
QTLRQVRANLSEPEPKLRQRLQEIYDNSDSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYSNYAAL
VSGRTFSTGDLADGNTDVFINIDLKTLETHAGLARVIIGSFLNGIYNRNGTMPGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGTTTVE
IDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGTN
RFHRLRGTPEASRIEPARSATSKADPEQ
MSRIMLFIAPVALMILVAAGLTGIEHWLSGLGKTEAARQTLGRAGIALPYVSAGLVGIVFLFASAGAIRI
KTAGWGVVAGGAATILIAAIRETMRLSALTDKVPAGMSIFSYVDPATLIGAAAAAIAACFALRVALAGNA
AFASAEPKRVRGKRALHGEAEWMKLPEAANLFSDTGGIVIGERYRVDKDSTAARSFRADEPETWGAGGKS
PLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWGGSLIVLDPSNEVAPMVSAHRTSANRDLFVLDPK
NPATGFNALDWIGKFGGTKEEDIASVASWIMSDSGGARGVRDDFFRASALQLLTAMIADVCLSGHTEKEN
QTLRQVRANLSEPEPKLRQRLQEIYDNSDSDFVKENVAAFVNMTPETFSGVYANAIKETHWLSYSNYAAL
VSGRTFSTGDLADGNTDVFINIDLKTLETHAGLARVIIGSFLNGIYNRNGTMPGRALFLLDEVARLGYMR
ILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWISFAAINDPETADYISRRCGTTTVE
IDQVSRSFQAKGSSRTRSKQLAARPLIQPHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVGTN
RFHRLRGTPEASRIEPARSATSKADPEQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 187156..194921
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MF410_RS00835 (MF410_00835) | 182204..182836 | - | 633 | WP_088395887 | DUF433 domain-containing protein | - |
| MF410_RS00840 (MF410_00840) | 182938..183159 | - | 222 | WP_088395888 | helix-turn-helix transcriptional regulator | - |
| MF410_RS00845 (MF410_00845) | 183388..183708 | + | 321 | WP_088395889 | transcriptional repressor TraM | - |
| MF410_RS00850 (MF410_00850) | 183712..184416 | - | 705 | WP_088395890 | autoinducer binding domain-containing protein | - |
| MF410_RS00855 (MF410_00855) | 184496..185236 | - | 741 | WP_096764645 | LuxR family transcriptional regulator | - |
| MF410_RS00860 (MF410_00860) | 185407..186701 | - | 1295 | Protein_172 | IncP-type conjugal transfer protein TrbI | - |
| MF410_RS00865 (MF410_00865) | 186713..187153 | - | 441 | WP_088395893 | conjugal transfer protein TrbH | - |
| MF410_RS00870 (MF410_00870) | 187156..187968 | - | 813 | WP_088395894 | P-type conjugative transfer protein TrbG | virB9 |
| MF410_RS00875 (MF410_00875) | 187986..188648 | - | 663 | WP_088395895 | conjugal transfer protein TrbF | virB8 |
| MF410_RS00880 (MF410_00880) | 188669..189850 | - | 1182 | WP_088395896 | P-type conjugative transfer protein TrbL | virB6 |
| MF410_RS00885 (MF410_00885) | 189844..190044 | - | 201 | WP_081284279 | entry exclusion protein TrbK | - |
| MF410_RS00890 (MF410_00890) | 190041..190844 | - | 804 | WP_064843797 | P-type conjugative transfer protein TrbJ | virB5 |
| MF410_RS00895 (MF410_00895) | 190816..193271 | - | 2456 | Protein_179 | conjugal transfer protein TrbE | - |
| MF410_RS00900 (MF410_00900) | 193282..193581 | - | 300 | WP_064843799 | conjugal transfer protein TrbD | virB3 |
| MF410_RS00905 (MF410_00905) | 193574..193954 | - | 381 | WP_064843801 | TrbC/VirB2 family protein | virB2 |
| MF410_RS00910 (MF410_00910) | 193944..194921 | - | 978 | WP_064843803 | P-type conjugative transfer ATPase TrbB | virB11 |
| MF410_RS00915 (MF410_00915) | 194932..195555 | - | 624 | WP_141399552 | acyl-homoserine-lactone synthase | - |
| MF410_RS00920 (MF410_00920) | 195932..197156 | + | 1225 | Protein_184 | plasmid partitioning protein RepA | - |
| MF410_RS00925 (MF410_00925) | 197153..198190 | + | 1038 | WP_064843806 | plasmid partitioning protein RepB | - |
| MF410_RS00930 (MF410_00930) | 198376..199590 | + | 1215 | WP_239619760 | plasmid replication protein RepC | - |
Host bacterium
| ID | 5315 | GenBank | NZ_CP092347 |
| Plasmid name | pRSpC104g | Incompatibility group | - |
| Plasmid size | 237978 bp | Coordinate of oriT [Strand] | 176070..176111 [+] |
| Host baterium | Rhizobium sp. C104 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |