Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104844 |
| Name | oriT_pEcl4-2 |
| Organism | Enterobacter hormaechei strain Eho-4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP047742 (83086..83209 [-], 124 nt) |
| oriT length | 124 nt |
| IRs (inverted repeats) | 92..99, 113..120 (ATAATGTA..TACATTAT) 90..95, 107..112 (AAATAA..TTATTT) 39..46, 49..56 (GCAAAAAC..GTTTTTGC) 3..10, 15..22 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 124 nt
GGTTGGTGGTTCTCACCACCAAAAGCACCACACACTACGCAAAAACAAGTTTTTGCTGATTTGCTATTTGAATCATTAACTTATGTTTTAAATAATGTATTTTAATTTATTTTACATTATAAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file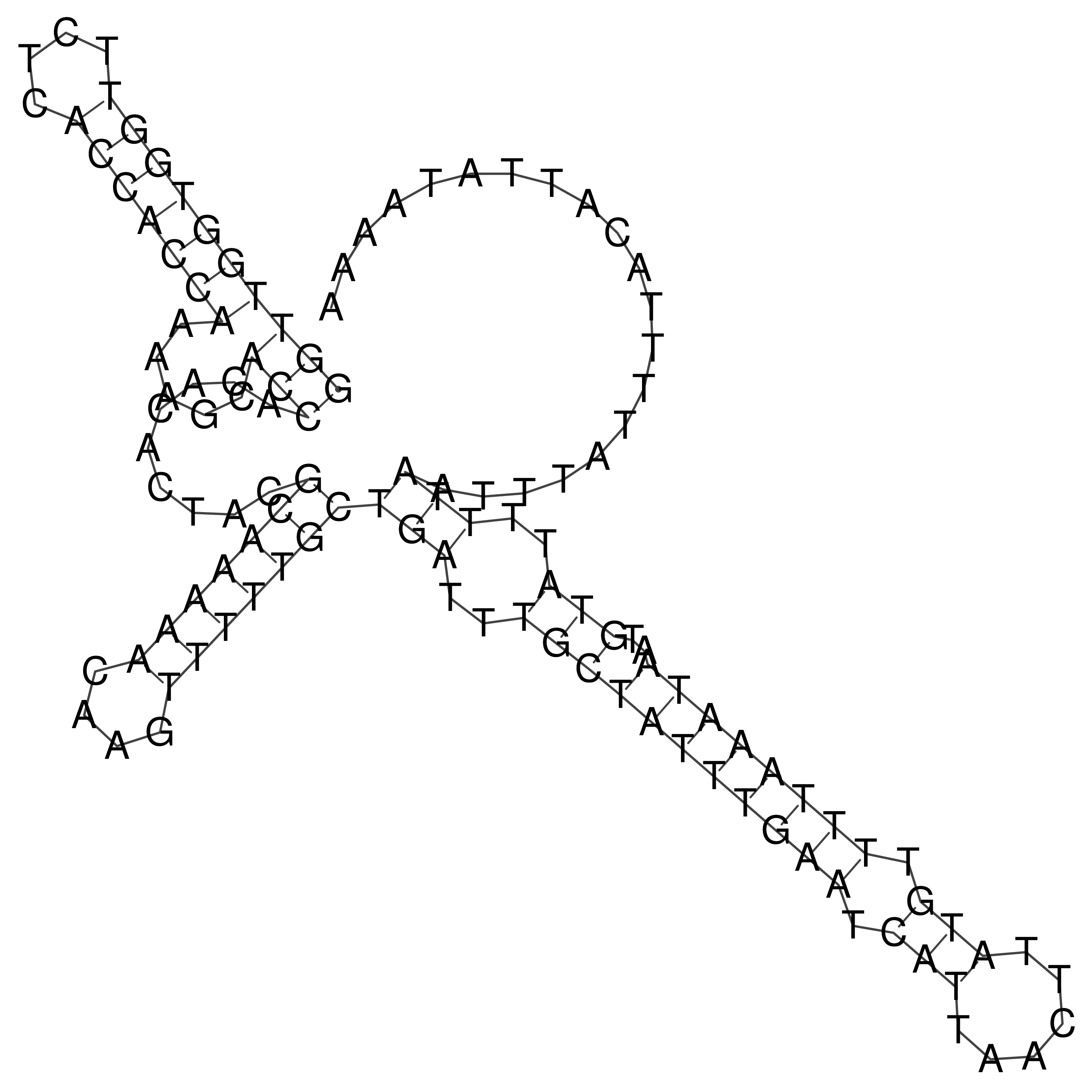
Relaxase
| ID | 3432 | GenBank | WP_000948349 |
| Name | traI_GVI65_RS25510_pEcl4-2 |
UniProt ID | C8CGS7 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191729.35 Da Isoelectric Point: 5.7956
MLSFSVVKSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQDPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFSENVLANRIAFGKIYQSELRQRVEA
LGYETEVVGKHGMWEMPGVPVEAFSSRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRAEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMVFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYDRLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETSLETAISLQKTGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGETFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWENNPDALALVDSVYHRIAGISKDDGLITLEDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAQRLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRSAISERETALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1523 | GenBank | WP_001254388 |
| Name | WP_001254388_pEcl4-2 |
UniProt ID | W9A0Z2 |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 9072.20 Da Isoelectric Point: 10.2071
MRRRNARGGISRTVSVYLDEDTNNRLIRAKDRSGRSKTIEVQIRLRDHLKRFPDFYNEEIFREVIEENES
TFKEL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | W9A0Z2 |
| ID | 1524 | GenBank | WP_001151562 |
| Name | WP_001151562_pEcl4-2 |
UniProt ID | A0A7I0KNG1 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14420.36 Da Isoelectric Point: 4.8624
MAKVQAYVSDEIVYKINKIVERRRAEGAKSTDVSFSSISTMLLELGLRVHEAQMERKESAFNQAEFNKVL
LECAVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIRDKVSSEMERFFPENDEE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0KNG1 |
T4CP
| ID | 3373 | GenBank | WP_000009347 |
| Name | traD_GVI65_RS25515_pEcl4-2 |
UniProt ID | _ |
| Length | 714 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 714 a.a. Molecular weight: 81087.57 Da Isoelectric Point: 5.1644
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLILWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGDPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPEVASGEDVTQAEQPQQPVSSVINDK
KSDAGVSVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPDIQQHMQRREEVNINV
HRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3374 | GenBank | WP_000069778 |
| Name | traC_GVI65_RS25615_pEcl4-2 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 99300.06 Da Isoelectric Point: 6.3461
MSNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAI
PINGANKSIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAA
ATQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASLQGASIATQTVDAQAFIDIV
GEMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLNVRADYLTLGLRENGRNSTARILNFHLARNPEIAF
LWNMADNYSNLLNPELSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWG
ELRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGK
GLFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFRGMNNTNYNMAVCGTSG
AGKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLS
VMASPNGNLDEVHEGLLLQAVRASWLAKENRARIDDVVDFLKNASDSEQYAGSPTIRSRLDEMIVLLDQY
TANGTYGRYFNSDEPSLRDDARMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRTLKKLNVIDEGW
RLLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKY
NQLFPDQFQPLQRDMIGKFGAAKDQWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREG
MSIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 55391..83781
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| GVI65_RS25515 (GVI65_25495) | 55391..57535 | - | 2145 | WP_000009347 | type IV conjugative transfer system coupling protein TraD | virb4 |
| GVI65_RS25520 (GVI65_25500) | 57586..58323 | - | 738 | WP_000199901 | hypothetical protein | - |
| GVI65_RS25525 (GVI65_25505) | 58526..59257 | - | 732 | WP_000782451 | conjugal transfer complement resistance protein TraT | - |
| GVI65_RS25530 (GVI65_25510) | 59306..59791 | - | 486 | WP_000605869 | hypothetical protein | - |
| GVI65_RS25535 (GVI65_25515) | 59807..62629 | - | 2823 | WP_001007080 | conjugal transfer mating-pair stabilization protein TraG | traG |
| GVI65_RS25540 (GVI65_25520) | 62626..63999 | - | 1374 | WP_000944331 | conjugal transfer pilus assembly protein TraH | traH |
| GVI65_RS25545 (GVI65_25525) | 63986..64378 | - | 393 | WP_000660700 | F-type conjugal transfer protein TrbF | - |
| GVI65_RS25550 (GVI65_25530) | 64359..64706 | - | 348 | WP_001309242 | P-type conjugative transfer protein TrbJ | - |
| GVI65_RS25555 (GVI65_25535) | 64636..65181 | - | 546 | WP_000059830 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| GVI65_RS25560 (GVI65_25540) | 65168..65452 | - | 285 | WP_000624107 | type-F conjugative transfer system pilin chaperone TraQ | - |
| GVI65_RS25565 (GVI65_25545) | 65533..65868 | + | 336 | WP_000736414 | hypothetical protein | - |
| GVI65_RS25570 (GVI65_25550) | 65849..66190 | - | 342 | WP_001298557 | conjugal transfer protein TrbA | - |
| GVI65_RS25575 (GVI65_25555) | 66204..66947 | - | 744 | WP_001030385 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| GVI65_RS25580 (GVI65_25560) | 66940..67197 | - | 258 | WP_000864314 | conjugal transfer protein TrbE | - |
| GVI65_RS25585 (GVI65_25565) | 67224..69074 | - | 1851 | WP_000821859 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| GVI65_RS25590 (GVI65_25570) | 69071..69709 | - | 639 | WP_001080251 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| GVI65_RS25595 (GVI65_25575) | 69718..70023 | - | 306 | WP_000224418 | hypothetical protein | - |
| GVI65_RS25600 (GVI65_25580) | 70053..71045 | - | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| GVI65_RS25605 (GVI65_25585) | 71042..71674 | - | 633 | WP_001203736 | type-F conjugative transfer system protein TraW | traW |
| GVI65_RS25610 (GVI65_25590) | 71671..72057 | - | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| GVI65_RS25615 (GVI65_25595) | 72054..74684 | - | 2631 | WP_000069778 | type IV secretion system protein TraC | virb4 |
| GVI65_RS27340 | 74810..75023 | - | 214 | Protein_81 | hypothetical protein | - |
| GVI65_RS25620 (GVI65_25600) | 75051..75269 | - | 219 | WP_000556746 | hypothetical protein | - |
| GVI65_RS25625 (GVI65_25605) | 75349..75822 | - | 474 | WP_000549566 | hypothetical protein | - |
| GVI65_RS25630 (GVI65_25610) | 75815..76036 | - | 222 | WP_001278694 | conjugal transfer protein TraR | - |
| GVI65_RS25635 (GVI65_25615) | 76171..76686 | - | 516 | WP_000809889 | type IV conjugative transfer system lipoprotein TraV | traV |
| GVI65_RS25640 (GVI65_25620) | 76683..76935 | - | 253 | Protein_86 | conjugal transfer protein TrbG | - |
| GVI65_RS25645 (GVI65_25625) | 76928..77248 | - | 321 | WP_001057274 | conjugal transfer protein TrbD | - |
| GVI65_RS25650 (GVI65_25630) | 77235..77825 | - | 591 | WP_000002787 | conjugal transfer pilus-stabilizing protein TraP | - |
| GVI65_RS25655 (GVI65_25635) | 77815..79242 | - | 1428 | WP_000146665 | F-type conjugal transfer pilus assembly protein TraB | traB |
| GVI65_RS25660 (GVI65_25640) | 79242..79970 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| GVI65_RS25665 (GVI65_25645) | 79957..80523 | - | 567 | WP_000399776 | type IV conjugative transfer system protein TraE | traE |
| GVI65_RS25670 (GVI65_25650) | 80545..80856 | - | 312 | WP_000012100 | type IV conjugative transfer system protein TraL | traL |
| GVI65_RS25675 (GVI65_25655) | 80871..81230 | - | 360 | WP_001098988 | type IV conjugative transfer system pilin TraA | - |
| GVI65_RS25680 (GVI65_25660) | 81264..81491 | - | 228 | WP_001254388 | conjugal transfer relaxosome protein TraY | - |
| GVI65_RS25685 (GVI65_25665) | 81585..82271 | - | 687 | WP_000332487 | PAS domain-containing protein | - |
| GVI65_RS25690 (GVI65_25670) | 82465..82848 | - | 384 | WP_001151562 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| GVI65_RS25695 (GVI65_25675) | 83191..83781 | + | 591 | WP_000252681 | transglycosylase SLT domain-containing protein | - |
| GVI65_RS25700 (GVI65_25680) | 84077..84898 | - | 822 | WP_001234489 | DUF932 domain-containing protein | - |
| GVI65_RS25705 (GVI65_25685) | 85017..85304 | - | 288 | WP_000107526 | hypothetical protein | - |
| GVI65_RS27195 | 85605..85902 | + | 298 | Protein_100 | hypothetical protein | - |
| GVI65_RS25710 (GVI65_25695) | 86220..86345 | - | 126 | WP_096937776 | type I toxin-antitoxin system Hok family toxin | - |
| GVI65_RS27200 | 86287..86436 | - | 150 | Protein_102 | DUF5431 family protein | - |
| GVI65_RS25715 (GVI65_25700) | 86610..87373 | - | 764 | Protein_103 | plasmid SOS inhibition protein A | - |
| GVI65_RS25720 (GVI65_25705) | 87370..87804 | - | 435 | WP_000845910 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 5282 | GenBank | NZ_CP047742 |
| Plasmid name | pEcl4-2 | Incompatibility group | IncFIB |
| Plasmid size | 114230 bp | Coordinate of oriT [Strand] | 83086..83209 [-] |
| Host baterium | Enterobacter hormaechei strain Eho-4 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | senB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |