Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104823 |
| Name | oriT_pUSP3 |
| Organism | Aminobacter sp. MSH1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP028971 (12953..12989 [+], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
>oriT_pUSP3
GGATCCAAGGGCGCAGTAATACGTCGCTGATGCGACG
GGATCCAAGGGCGCAGTAATACGTCGCTGATGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file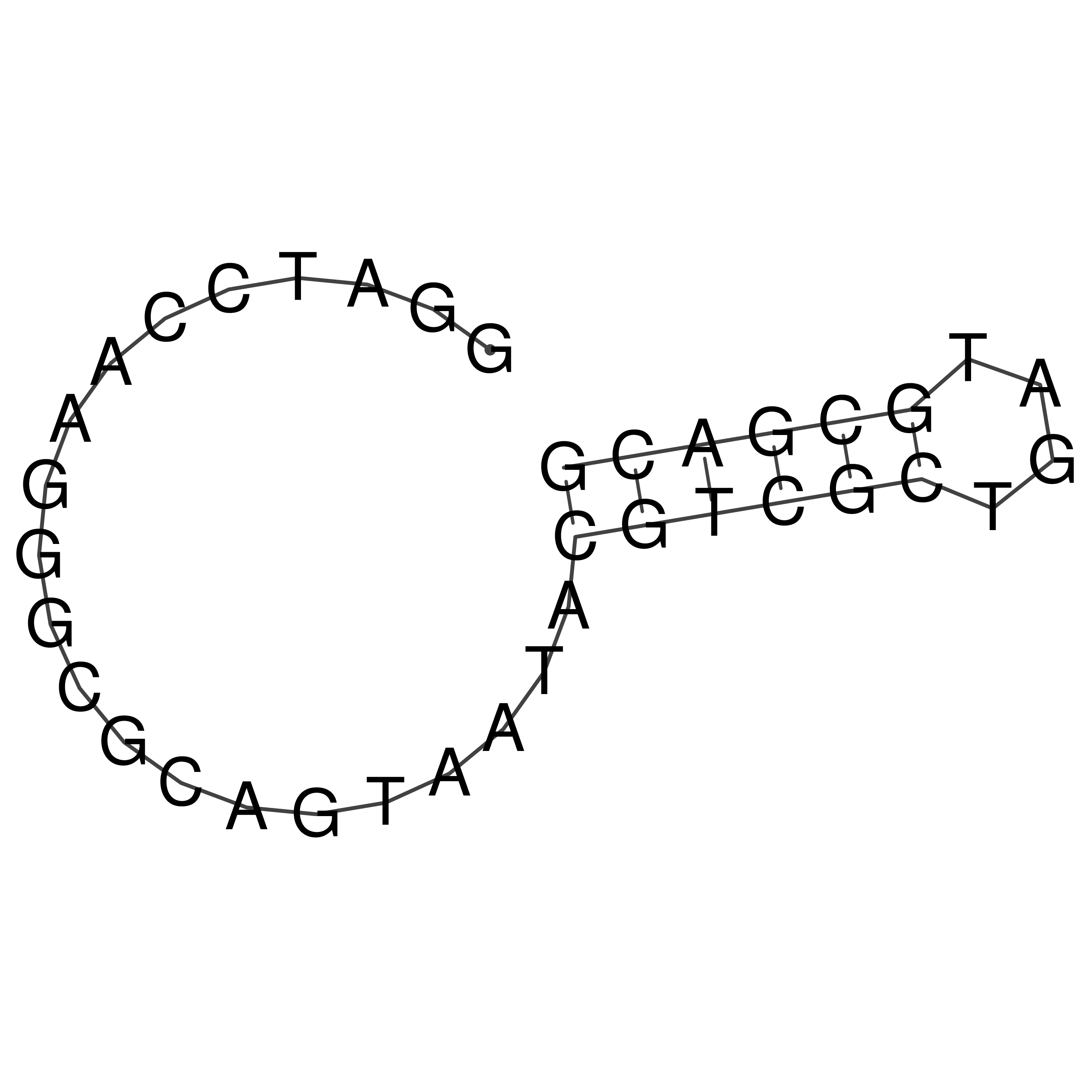
Relaxase
| ID | 3420 | GenBank | WP_108610784 |
| Name | traA_DCS14_RS30395_pUSP3 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123759.27 Da Isoelectric Point: 10.0608
>WP_108610784.1 Ti-type conjugative transfer relaxase TraA [Aminobacter sp. MSH1]
MAITHFTPQLIGRGNGRSAVLSAAYRHCAKMTYEMEARTVDYSNKRNLAYEEFLLPADAPDWARKLIADR
SVSGAVEAFWNRVEAFEKREDAQFAKEFIIALPAELTKEQNIALMRQFVFEQVLARGQVADWVYHDEPGN
PHVHLMTTLRPLTESGFGAKKVAVLGQDGQPVRTKTGKIQYRLWAGEKQDFLEQRNSWLDLQNQHLALGG
VEIRVDGRSYAERGIDLVPTVHIGVGAKAIQRKGVQQGWTPSLDRIEQFEAQRQENARRILRRPEIVLDL
LSSEKSVFDSQDIAKVLHRYVDDAGTFQHLMTRIVQSPDLLRIEREGVEFATGERVRARYTTRELIRVEA
EMAKRAIWLSERTSHGVRQKVLDATFARHERLSQEQRVAIEHVAGAARIAAVVGRAGAGKTTMMKAAREA
WESAGYRVVGGALAGKAAEGLEKEAGIASRTLASWELRWKEGRDALDGKTVFVLDEAGMVASKQMALFVE
AALRTGAKLVLVGDPEQLQPIEAGAAFRAIVERIGYAELETIYRQREDWMRKASLDLARSRVDQAISAYR
GRGRVMGSDLKAQAVENLIADWNSDYDATKSTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFRTEDG
MRRFDAGDQIVFLKNDGSLGVKNGMIAKVVEVAPNRIVAEIGEGDQRHRVSVDQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVALTRHREDMALYYGRRSFGMNGGLVKVLSRRNAKETTLDYERGTLY
RQALRFAENRGLHIARVARTLVRDRVDWTLRQKQRLADLGLRLRGFGERLGLLDTPQTQTLKECAPMVKG
VTQFPYSVQDTVEEKLLADPAVKKQWEEVSTRFRFVFADPETAFRAMNFDAVLTDPSTAKATLERLVSEP
ATIGPLKGRVGLLAGKADREDRHIAQVTAPALKRDIERYLRLRQTAVQKVEAEEQTLRRRFSIDIPALSP
AAIQVLERVRDAIDRNDLPTALGYALADRMAKAEIDGFNKAITERFGERTLLTNAAREPSGKLFDRAAEG
LAPDERQKLAEAWPVMRAAQQIAAHERTVETLKQAENLRLSQRQTPVLKQ
MAITHFTPQLIGRGNGRSAVLSAAYRHCAKMTYEMEARTVDYSNKRNLAYEEFLLPADAPDWARKLIADR
SVSGAVEAFWNRVEAFEKREDAQFAKEFIIALPAELTKEQNIALMRQFVFEQVLARGQVADWVYHDEPGN
PHVHLMTTLRPLTESGFGAKKVAVLGQDGQPVRTKTGKIQYRLWAGEKQDFLEQRNSWLDLQNQHLALGG
VEIRVDGRSYAERGIDLVPTVHIGVGAKAIQRKGVQQGWTPSLDRIEQFEAQRQENARRILRRPEIVLDL
LSSEKSVFDSQDIAKVLHRYVDDAGTFQHLMTRIVQSPDLLRIEREGVEFATGERVRARYTTRELIRVEA
EMAKRAIWLSERTSHGVRQKVLDATFARHERLSQEQRVAIEHVAGAARIAAVVGRAGAGKTTMMKAAREA
WESAGYRVVGGALAGKAAEGLEKEAGIASRTLASWELRWKEGRDALDGKTVFVLDEAGMVASKQMALFVE
AALRTGAKLVLVGDPEQLQPIEAGAAFRAIVERIGYAELETIYRQREDWMRKASLDLARSRVDQAISAYR
GRGRVMGSDLKAQAVENLIADWNSDYDATKSTLILAHLRRDVRMLNVMAREKLVERGIVGEGHVFRTEDG
MRRFDAGDQIVFLKNDGSLGVKNGMIAKVVEVAPNRIVAEIGEGDQRHRVSVDQRFYRNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVALTRHREDMALYYGRRSFGMNGGLVKVLSRRNAKETTLDYERGTLY
RQALRFAENRGLHIARVARTLVRDRVDWTLRQKQRLADLGLRLRGFGERLGLLDTPQTQTLKECAPMVKG
VTQFPYSVQDTVEEKLLADPAVKKQWEEVSTRFRFVFADPETAFRAMNFDAVLTDPSTAKATLERLVSEP
ATIGPLKGRVGLLAGKADREDRHIAQVTAPALKRDIERYLRLRQTAVQKVEAEEQTLRRRFSIDIPALSP
AAIQVLERVRDAIDRNDLPTALGYALADRMAKAEIDGFNKAITERFGERTLLTNAAREPSGKLFDRAAEG
LAPDERQKLAEAWPVMRAAQQIAAHERTVETLKQAENLRLSQRQTPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3358 | GenBank | WP_108610716 |
| Name | traG_DCS14_RS30380_pUSP3 |
UniProt ID | _ |
| Length | 641 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 641 a.a. Molecular weight: 68781.66 Da Isoelectric Point: 9.1176
>WP_108610716.1 Ti-type conjugative transfer system protein TraG [Aminobacter sp. MSH1]
MTANKLLLAGSAAALMLGACIGISGVETWLAGFGTSDAARLMLGRTGIALPYIAAAVIGVIFLFASAGAS
AIRAAGWGVAAGTVGVVAIAIAREAVRLSALADRVPAGRTILSYVDPATMAGATIAMICGGFAMRVAMRG
NAAFAVATPRRIRGKRAVHGEADWMSLPAAARLFPDSGGIAIGERYRVDRDSVAAEPFRADARETWGGGG
RSPLICFDGSFGSSHGIVFAGSGGFKTTSITVPTALKWGGSLVVLDPSSEVAPMVIGHRRRAGRNVFLLD
PGNAAIGFNALDWIGRFGSTKEEDIVAVATWIITDNPRQASARDDFFRASALQLLTVLIADVCLSGHTEE
KDQTLRRVRSNLSEPEPKLRERLTAIHDQSESAFVKENVAPFIAMTPETFSGVYANAVKETHWLSYSNYA
ALVSGSSFTTDELAAGETDIFIALDLKVLETHPGLARVIIGALMNAIYNRNGEVKGRTLFLLDEVARLGF
LRVLETARDAGRKYGITMTLLYQSIGQMREAYGGRDATSKWFESASWVSFAAINDPETADYISKRCGDTT
VEVDQTSRSSQMSGSSRTRSKQLTRRPLILPHEVMQMRADEQIVFTAGNPPLRCGRAIWFRRADMKACVG
ENRFHRKETEA
MTANKLLLAGSAAALMLGACIGISGVETWLAGFGTSDAARLMLGRTGIALPYIAAAVIGVIFLFASAGAS
AIRAAGWGVAAGTVGVVAIAIAREAVRLSALADRVPAGRTILSYVDPATMAGATIAMICGGFAMRVAMRG
NAAFAVATPRRIRGKRAVHGEADWMSLPAAARLFPDSGGIAIGERYRVDRDSVAAEPFRADARETWGGGG
RSPLICFDGSFGSSHGIVFAGSGGFKTTSITVPTALKWGGSLVVLDPSSEVAPMVIGHRRRAGRNVFLLD
PGNAAIGFNALDWIGRFGSTKEEDIVAVATWIITDNPRQASARDDFFRASALQLLTVLIADVCLSGHTEE
KDQTLRRVRSNLSEPEPKLRERLTAIHDQSESAFVKENVAPFIAMTPETFSGVYANAVKETHWLSYSNYA
ALVSGSSFTTDELAAGETDIFIALDLKVLETHPGLARVIIGALMNAIYNRNGEVKGRTLFLLDEVARLGF
LRVLETARDAGRKYGITMTLLYQSIGQMREAYGGRDATSKWFESASWVSFAAINDPETADYISKRCGDTT
VEVDQTSRSSQMSGSSRTRSKQLTRRPLILPHEVMQMRADEQIVFTAGNPPLRCGRAIWFRRADMKACVG
ENRFHRKETEA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3359 | GenBank | WP_108610730 |
| Name | t4cp2_DCS14_RS30470_pUSP3 |
UniProt ID | _ |
| Length | 818 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 818 a.a. Molecular weight: 91435.78 Da Isoelectric Point: 5.6624
>WP_108610730.1 conjugal transfer protein TrbE [Aminobacter sp. MSH1]
MAALRSFRHSGPSFSDLVPYAALVANGVILLKDGSLMAGWYFAGPDSESSTDTERNEVSRQINTVLSRLG
SGWMIQVEAVRVGTSDYPANDQCHFPDPVTLAIDGERRTHFQKERGHYESRHALILTWRPPEKTRSGLAR
YVYSDSESRTARYADTVLDSFQTSIREVEQYLGNVLSIRRMVTQSVEERGGFQTARYDELFQFIRFCITG
ENHPIRLPEIPMYLDWLATAEFHHGLSPMVENHYLAVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAQASSQLVAYGYYTPVIVL
FDDSEARLREKAEAVRRLIQAEGFGARIETVNATEAFLGSLPGNWYANIREPLINTRNLADLIPLNSVWS
GAPAAPCPFYPPASPPLMLVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLVAAQFRRYAFAQIFA
FDKGRSMLPLTLAAGGDHYEIGETEGEGGPRLAFCPLSDLATDGDRAWASEWIETLVVLQGVNITPDHRN
AISRQVALMADSSGRSLSDFVSGVQLREIKEALHHYTVDGPMGQLLDAESDGLSLGSFQTFEIEQLMNMG
ERSLVPVLLYLFRRIEKRLTGAPSLIILDEAWLMLGHATFRDKIREWLKVLRKANCAVVLATQSITDAER
SGIIDVLKESCPTKICLPNGAARERGTREFYERIGFNERQIEIVATALPKREYYVVSPDGRRLFDMALGP
VALAFAGASGKEDLKRIQALSAEHGGDWPVHWLQQRGIGDAETILERG
MAALRSFRHSGPSFSDLVPYAALVANGVILLKDGSLMAGWYFAGPDSESSTDTERNEVSRQINTVLSRLG
SGWMIQVEAVRVGTSDYPANDQCHFPDPVTLAIDGERRTHFQKERGHYESRHALILTWRPPEKTRSGLAR
YVYSDSESRTARYADTVLDSFQTSIREVEQYLGNVLSIRRMVTQSVEERGGFQTARYDELFQFIRFCITG
ENHPIRLPEIPMYLDWLATAEFHHGLSPMVENHYLAVVAIDGFPAESWPGILNSLDLMPLTYRWSSRFIF
LDDQEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMMMVAETEDAIAQASSQLVAYGYYTPVIVL
FDDSEARLREKAEAVRRLIQAEGFGARIETVNATEAFLGSLPGNWYANIREPLINTRNLADLIPLNSVWS
GAPAAPCPFYPPASPPLMLVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLVAAQFRRYAFAQIFA
FDKGRSMLPLTLAAGGDHYEIGETEGEGGPRLAFCPLSDLATDGDRAWASEWIETLVVLQGVNITPDHRN
AISRQVALMADSSGRSLSDFVSGVQLREIKEALHHYTVDGPMGQLLDAESDGLSLGSFQTFEIEQLMNMG
ERSLVPVLLYLFRRIEKRLTGAPSLIILDEAWLMLGHATFRDKIREWLKVLRKANCAVVLATQSITDAER
SGIIDVLKESCPTKICLPNGAARERGTREFYERIGFNERQIEIVATALPKREYYVVSPDGRRLFDMALGP
VALAFAGASGKEDLKRIQALSAEHGGDWPVHWLQQRGIGDAETILERG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 10389..32859
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DCS14_RS30345 | 6005..6589 | + | 585 | WP_108610712 | hypothetical protein | - |
| DCS14_RS30350 | 6910..7836 | + | 927 | WP_108610713 | zincin-like metallopeptidase domain-containing protein | - |
| DCS14_RS30355 | 7836..8015 | + | 180 | Protein_11 | hypothetical protein | - |
| DCS14_RS30365 | 8632..8946 | + | 315 | WP_108610714 | DUF736 domain-containing protein | - |
| DCS14_RS31760 | 9028..9174 | + | 147 | WP_159097927 | hypothetical protein | - |
| DCS14_RS30370 | 9263..9508 | - | 246 | WP_108610715 | WGR domain-containing protein | - |
| DCS14_RS30375 | 9593..9979 | - | 387 | WP_108610783 | thermonuclease family protein | - |
| DCS14_RS30380 | 10389..12314 | - | 1926 | WP_108610716 | Ti-type conjugative transfer system protein TraG | virb4 |
| DCS14_RS30385 | 12301..12516 | - | 216 | WP_108610717 | type IV conjugative transfer system coupling protein TraD | - |
| DCS14_RS30390 | 12521..12817 | - | 297 | WP_108610718 | conjugal transfer protein TraC | - |
| DCS14_RS30395 | 13066..16368 | + | 3303 | WP_108610784 | Ti-type conjugative transfer relaxase TraA | - |
| DCS14_RS30400 | 16365..16895 | + | 531 | WP_108610719 | conjugative transfer signal peptidase TraF | - |
| DCS14_RS30405 | 16912..18051 | + | 1140 | WP_245453616 | conjugal transfer protein TraB | - |
| DCS14_RS30410 | 18066..18680 | + | 615 | WP_108610721 | TraH family protein | virB1 |
| DCS14_RS30415 | 18971..19294 | + | 324 | WP_108610722 | transcriptional repressor TraM | - |
| DCS14_RS30420 | 19309..20013 | - | 705 | WP_245453614 | autoinducer binding domain-containing protein | - |
| DCS14_RS30425 | 20398..22011 | + | 1614 | WP_108610723 | hypothetical protein | - |
| DCS14_RS30430 | 22001..23026 | + | 1026 | WP_108610724 | dienelactone hydrolase-related enzyme | - |
| DCS14_RS30435 | 23290..24594 | - | 1305 | WP_108610725 | IncP-type conjugal transfer protein TrbI | virB10 |
| DCS14_RS30440 | 24606..25058 | - | 453 | WP_108610726 | conjugal transfer protein TrbH | - |
| DCS14_RS30445 | 25062..25901 | - | 840 | WP_108610786 | P-type conjugative transfer protein TrbG | virB9 |
| DCS14_RS30450 | 25919..26581 | - | 663 | WP_108610727 | conjugal transfer protein TrbF | virB8 |
| DCS14_RS30455 | 26600..27787 | - | 1188 | WP_108610728 | P-type conjugative transfer protein TrbL | virB6 |
| DCS14_RS30460 | 27781..27990 | - | 210 | WP_245453615 | entry exclusion protein TrbK | - |
| DCS14_RS30465 | 27987..28787 | - | 801 | WP_108610729 | P-type conjugative transfer protein TrbJ | virB5 |
| DCS14_RS30470 | 28759..31215 | - | 2457 | WP_108610730 | conjugal transfer protein TrbE | virb4 |
| DCS14_RS30475 | 31226..31537 | - | 312 | WP_108610731 | conjugal transfer protein TrbD | virB3 |
| DCS14_RS30480 | 31530..31901 | - | 372 | WP_108610732 | TrbC/VirB2 family protein | virB2 |
| DCS14_RS30485 | 31891..32859 | - | 969 | WP_108610733 | P-type conjugative transfer ATPase TrbB | virB11 |
| DCS14_RS30490 | 33421..34614 | + | 1194 | WP_067970026 | plasmid partitioning protein RepA | - |
| DCS14_RS30495 | 34836..35759 | + | 924 | WP_343207477 | plasmid partitioning protein RepB | - |
| DCS14_RS30500 | 35934..37277 | + | 1344 | WP_108610735 | plasmid replication protein RepC | - |
| DCS14_RS30505 | 37327..37854 | - | 528 | Protein_41 | recombinase family protein | - |
Host bacterium
| ID | 5261 | GenBank | NZ_CP028971 |
| Plasmid name | pUSP3 | Incompatibility group | - |
| Plasmid size | 97029 bp | Coordinate of oriT [Strand] | 12953..12989 [+] |
| Host baterium | Aminobacter sp. MSH1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |