Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104819 |
| Name | oriT_pAM01 |
| Organism | Aminobacter sp. MSH1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP026266 (64214..64250 [+], 37 nt) |
| oriT length | 37 nt |
| IRs (inverted repeats) | 22..27, 32..37 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 37 nt
>oriT_pAM01
GGGTCCAAGGGCGCAATTATACGTCGCTGACGCGACG
GGGTCCAAGGGCGCAATTATACGTCGCTGACGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file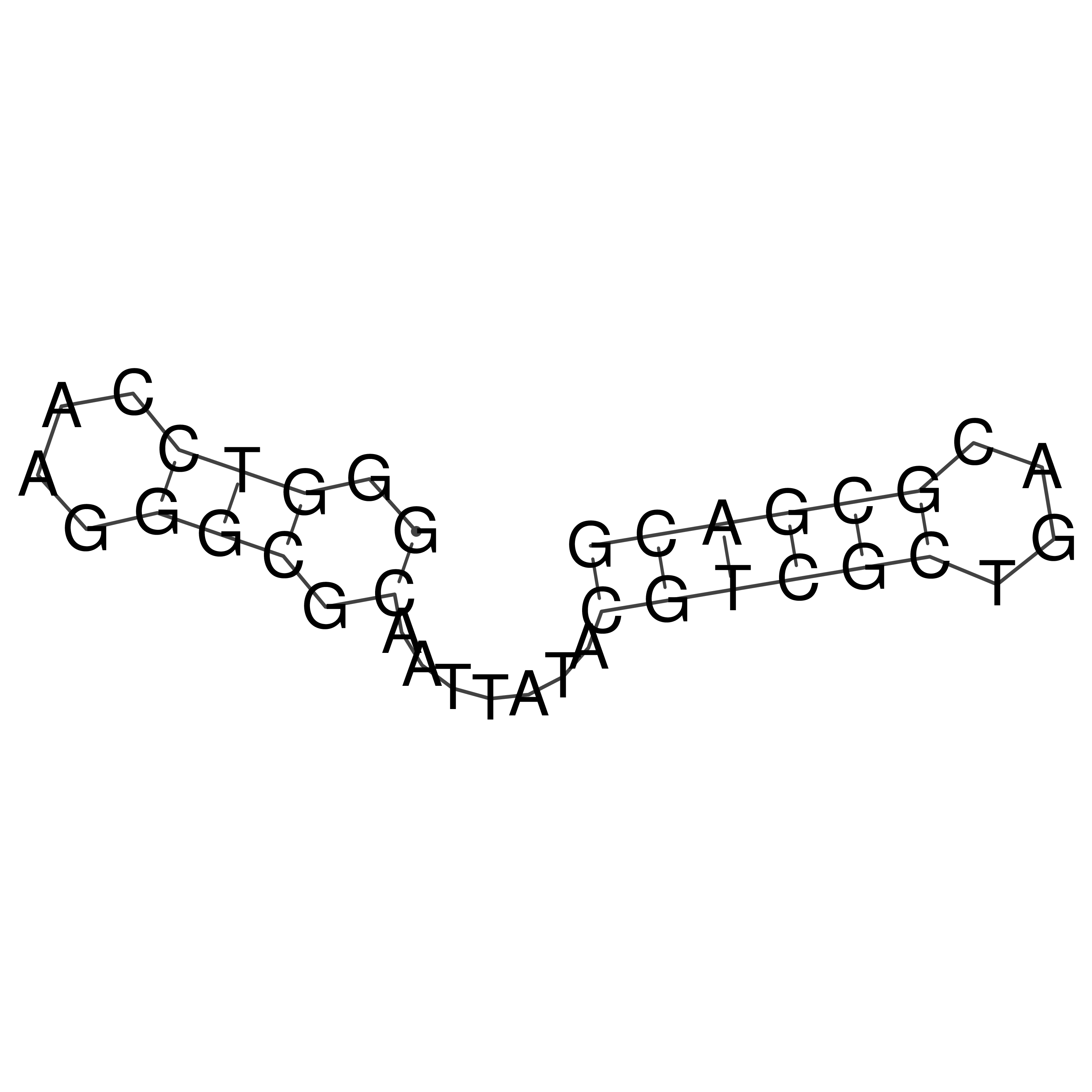
Relaxase
| ID | 3416 | GenBank | WP_108610369 |
| Name | traA_CO731_RS26705_pAM01 |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 122823.15 Da Isoelectric Point: 10.0350
>WP_108610369.1 Ti-type conjugative transfer relaxase TraA [Aminobacter sp. MSH1]
MAVPHFSVAIISRGNGRSAVLSAAYRHCAKMEYEREARSIDYTRKQGLLHEEFLAPPDAPQWVSSLIADR
SVSGAAEAFWNKVEAFEKRSDAQLAKDITMALPLELSAKQNIALVRDFVEQHILAKGMVADWVYHDAPGN
PHVHLMMTLRPLTEDGFGAKKIAVIGENGEVLRNQAGKIVYELWAGGVADFTAFRDAWFTTQNKHLALGG
IDLRVDGRSYAERGIDLVPIIHVGVGAQAIQRKSVSDGSQPSLERIELFEARRKENAGRILKRPEIVLEM
ISSEKSVFDERDIAKILHRYVDDAGTFQHLMVRILQSPEVLRIERNGIDLATGERLRSRYTTRELIRLEA
RMAKQSLRLLQRATHGVQEKVLDSVFARHSRLSQEQRVAIEHVAGAGKIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGAALAGKAAEGLEKEAGIVSRTLASWELRWQEGRDLLDARTVFVLDEAGMVASKQMALVVE
AATRAGAKLVLIGDPDQLQPIEAGAAFRAIADRIGYAELETIYRQRDDWMRAASLDLARGRVDEAVSAYR
QHGRVLGSELKAQAIDSLIADWNRDYDPAETSLILAHLRRDVRILNDMARERLVERGIVGEGHAFRTEDG
LRQFDAGDQIVFLKNETSLGVKNGMIGRVMDAAANRIVAAIGEGEHRRLVTVEQRFYHNIDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDMSLYYGRRSFGLNGGLIAILARRNAKETTLDYEHGTLY
RQALRFAENRGLNFVRIARTLVRDRLDWTLRQRQRLADLTQQMRAHGERLGLLQPPSTQTWKAAEPMVAG
VTLFARSLADVVEERLLADPALAKQWDEASTRFRYIFTDPETAFRSMNFDAVLTDAKVAKQTLDRLVSEP
GSIGPLKGRAGILASRADKEARRVAEVNVPALKRDIERYLQLRQTAAERIEREEQSLRHRVSIDIPALSL
AARSVLEKIRDAIARNDLAAARGHALSSREAKLEIDGFNRAVTERFGERALLANTAREPSGTLFDKAAAG
LRPGEREKLHQAWPVMRAAQQIAAHERTAQTLKQAEHLRLAQRQSTAWKQ
MAVPHFSVAIISRGNGRSAVLSAAYRHCAKMEYEREARSIDYTRKQGLLHEEFLAPPDAPQWVSSLIADR
SVSGAAEAFWNKVEAFEKRSDAQLAKDITMALPLELSAKQNIALVRDFVEQHILAKGMVADWVYHDAPGN
PHVHLMMTLRPLTEDGFGAKKIAVIGENGEVLRNQAGKIVYELWAGGVADFTAFRDAWFTTQNKHLALGG
IDLRVDGRSYAERGIDLVPIIHVGVGAQAIQRKSVSDGSQPSLERIELFEARRKENAGRILKRPEIVLEM
ISSEKSVFDERDIAKILHRYVDDAGTFQHLMVRILQSPEVLRIERNGIDLATGERLRSRYTTRELIRLEA
RMAKQSLRLLQRATHGVQEKVLDSVFARHSRLSQEQRVAIEHVAGAGKIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGAALAGKAAEGLEKEAGIVSRTLASWELRWQEGRDLLDARTVFVLDEAGMVASKQMALVVE
AATRAGAKLVLIGDPDQLQPIEAGAAFRAIADRIGYAELETIYRQRDDWMRAASLDLARGRVDEAVSAYR
QHGRVLGSELKAQAIDSLIADWNRDYDPAETSLILAHLRRDVRILNDMARERLVERGIVGEGHAFRTEDG
LRQFDAGDQIVFLKNETSLGVKNGMIGRVMDAAANRIVAAIGEGEHRRLVTVEQRFYHNIDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDMSLYYGRRSFGLNGGLIAILARRNAKETTLDYEHGTLY
RQALRFAENRGLNFVRIARTLVRDRLDWTLRQRQRLADLTQQMRAHGERLGLLQPPSTQTWKAAEPMVAG
VTLFARSLADVVEERLLADPALAKQWDEASTRFRYIFTDPETAFRSMNFDAVLTDAKVAKQTLDRLVSEP
GSIGPLKGRAGILASRADKEARRVAEVNVPALKRDIERYLQLRQTAAERIEREEQSLRHRVSIDIPALSL
AARSVLEKIRDAIARNDLAAARGHALSSREAKLEIDGFNRAVTERFGERALLANTAREPSGTLFDKAAAG
LRPGEREKLHQAWPVMRAAQQIAAHERTAQTLKQAEHLRLAQRQSTAWKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 5257 | GenBank | NZ_CP026266 |
| Plasmid name | pAM01 | Incompatibility group | - |
| Plasmid size | 367312 bp | Coordinate of oriT [Strand] | 64214..64250 [+] |
| Host baterium | Aminobacter sp. MSH1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |