Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104805 |
| Name | oriT_pP13_h |
| Organism | Phaeobacter piscinae strain P13 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP010775 (33816..33835 [+], 20 nt) |
| oriT length | 20 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 20 nt
>oriT_pP13_h
GTTTGCATAAGTGCGCCCTT
GTTTGCATAAGTGCGCCCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file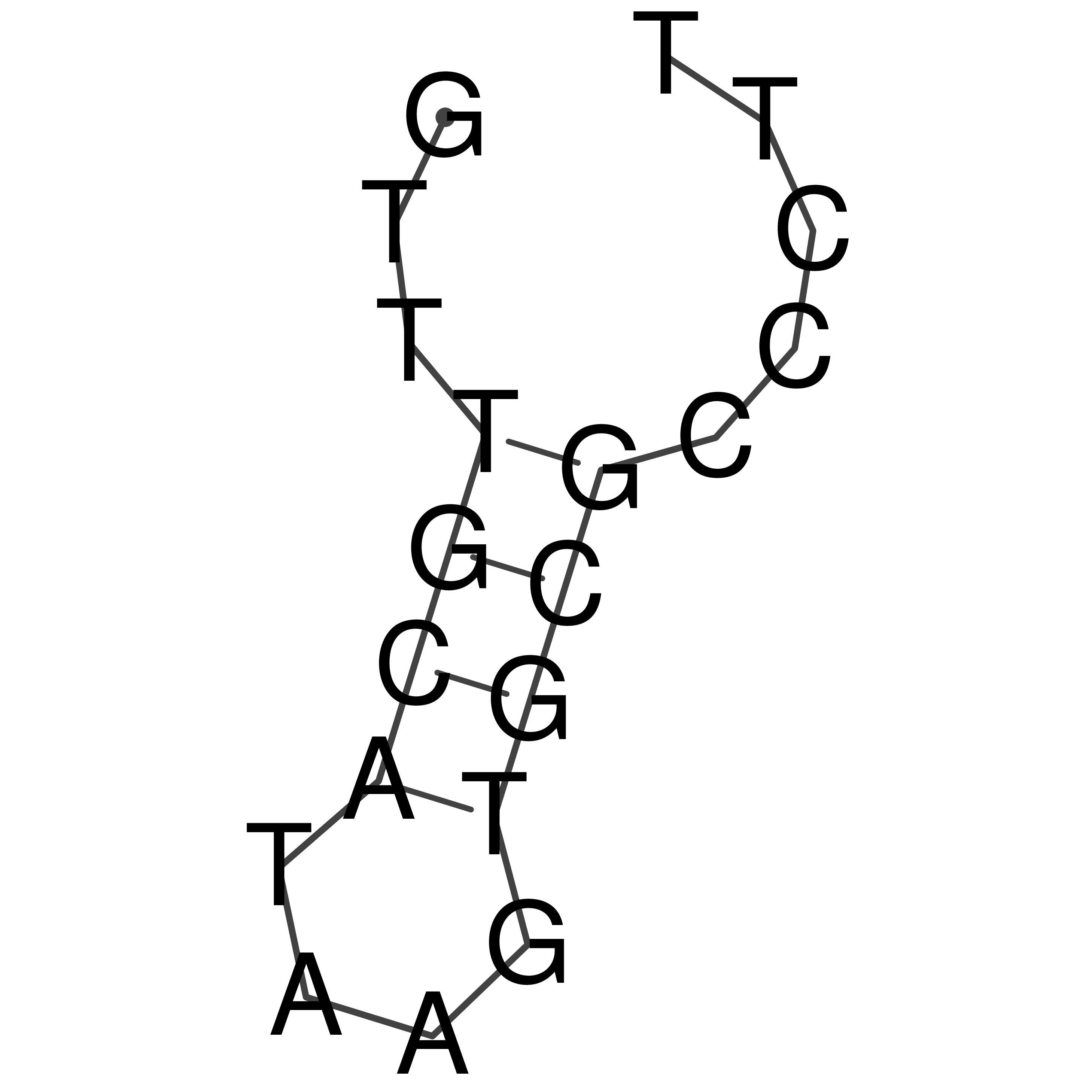
Relaxase
| ID | 3407 | GenBank | WP_067267002 |
| Name | mobQ_PhaeoP13_RS20835_pP13_h |
UniProt ID | A0A167GVD0 |
| Length | 430 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 430 a.a. Molecular weight: 48835.92 Da Isoelectric Point: 6.9896
>WP_067267002.1 MULTISPECIES: MobQ family relaxase [Roseobacteraceae]
MASYHLSVKTIKRSAGRSATAAAAYRVGERIECQREGRVHDYTRKQGIEETFILAPKDAPDWATDRSRLW
NEVEARETRRNSVTAREWELALPSEISAEDRSQITRDFAQELVSRYGVAVDVAIHAPHREGDQRNHHAHV
LTSTRRLEPEGFTAKTRVLDSAKTGGVEIEQMRGLWAELQNRALERAGEVERVDHRSLEAQREAAQERGD
KLSAEELDRDPELKLGPAANSMERRAKAMAERQGREYKPVTERGAVVHAARQARAAFREMRARLELARET
YGIEREAGQGRVSAGLAALRAATAKDHDRDRNPDDFRERLARVLGRSRDQDDTPKPEGRNYARERLKEIM
EKDAGRDGQAAVHKLDGHSEYDLGEETGRAARKPSVNERLKDVLNKPRGRLEIEDEREQERGDDENENTR
DKDRGEGHSL
MASYHLSVKTIKRSAGRSATAAAAYRVGERIECQREGRVHDYTRKQGIEETFILAPKDAPDWATDRSRLW
NEVEARETRRNSVTAREWELALPSEISAEDRSQITRDFAQELVSRYGVAVDVAIHAPHREGDQRNHHAHV
LTSTRRLEPEGFTAKTRVLDSAKTGGVEIEQMRGLWAELQNRALERAGEVERVDHRSLEAQREAAQERGD
KLSAEELDRDPELKLGPAANSMERRAKAMAERQGREYKPVTERGAVVHAARQARAAFREMRARLELARET
YGIEREAGQGRVSAGLAALRAATAKDHDRDRNPDDFRERLARVLGRSRDQDDTPKPEGRNYARERLKEIM
EKDAGRDGQAAVHKLDGHSEYDLGEETGRAARKPSVNERLKDVLNKPRGRLEIEDEREQERGDDENENTR
DKDRGEGHSL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A167GVD0 |
T4CP
| ID | 3343 | GenBank | WP_071974173 |
| Name | t4cp2_PhaeoP13_RS20770_pP13_h |
UniProt ID | _ |
| Length | 309 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 309 a.a. Molecular weight: 34655.54 Da Isoelectric Point: 5.4963
>WP_071974173.1 MULTISPECIES: nucleotidyl transferase AbiEii/AbiGii toxin family protein [Roseobacteraceae]
MSDNDPHAQDRTFDIVDVDVRAWVETARANPTLYRDRQVTEIVLGAIGLAPSLSKTLVLKGGAVMALAFK
SNRVTGDVDFTSMAEPADLTEKITAELNEILPSTAIKLGYLDLICRVQSVKKMPRAENFEDHEFPALKVK
VGSATRGTAEAARLADGKASRVLDVEISFRDQVYAFQELNLHGAGVAVRAFTIHELIAEKFRALLQQITR
KHKRNRRQDVYDIAFLIDEHDFGDADKTAILTTFIEKCRSRGIEVKPDSMDDPEIKQRAQADWETLALEI
GDLPPFEERFALMRDLYVSLPWGGIKKDR
MSDNDPHAQDRTFDIVDVDVRAWVETARANPTLYRDRQVTEIVLGAIGLAPSLSKTLVLKGGAVMALAFK
SNRVTGDVDFTSMAEPADLTEKITAELNEILPSTAIKLGYLDLICRVQSVKKMPRAENFEDHEFPALKVK
VGSATRGTAEAARLADGKASRVLDVEISFRDQVYAFQELNLHGAGVAVRAFTIHELIAEKFRALLQQITR
KHKRNRRQDVYDIAFLIDEHDFGDADKTAILTTFIEKCRSRGIEVKPDSMDDPEIKQRAQADWETLALEI
GDLPPFEERFALMRDLYVSLPWGGIKKDR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3344 | GenBank | WP_096873673 |
| Name | t4cp2_PhaeoP13_RS20850_pP13_h |
UniProt ID | _ |
| Length | 556 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 556 a.a. Molecular weight: 61380.31 Da Isoelectric Point: 6.8841
>WP_096873673.1 type IV secretory system conjugative DNA transfer family protein [Phaeobacter piscinae]
MRSVMLVFGGLFRFFGRLIFTPIMLGWLIGAMAFGAMLGTLVATPFIFTFFDQTPGESAWQWLVFGPFMF
VGAVFGFQYWRMASGADAYFGLTGDSHGSARFANRKELKKLERSKGQDDAGLLIGRNPHTGRLLRYDGPA
HLITLAPTRAGKGVGTVIPNLLAAERSVLVIDPKGENARIAGEARRRFGTVHVLDPFEVSGHPSACYNPL
DRLTPHSLDLGEDAASLTEALVMDPPGQVTEAHWNEEAKAILGGLIMFCVCHEDRDRRSLATVREYLTLP
PEKLRALLELMQDSDEAGGLIARAANRFLGKADREAASVLSNAQRHTHFLDSPRIAKCLARSDFAFSDLR
HRITSVFLVLPPNRMDAYSRWLRLLVSQALQDIARDAERPQNASEGRSERLTAPALFLLDEFAALGRLEA
VERAMGLMAGYGLQLWPILQDMSQLKDLYGERAGTFIANAGVQQVFGVNDFETAKWLSQMIGQETAGFQT
DSFKPGDGPSFSNNLTGRDLLTPDEIMQLPPKRQLLRVQGQATAVAQKLRYYADPEFAGLFTPETP
MRSVMLVFGGLFRFFGRLIFTPIMLGWLIGAMAFGAMLGTLVATPFIFTFFDQTPGESAWQWLVFGPFMF
VGAVFGFQYWRMASGADAYFGLTGDSHGSARFANRKELKKLERSKGQDDAGLLIGRNPHTGRLLRYDGPA
HLITLAPTRAGKGVGTVIPNLLAAERSVLVIDPKGENARIAGEARRRFGTVHVLDPFEVSGHPSACYNPL
DRLTPHSLDLGEDAASLTEALVMDPPGQVTEAHWNEEAKAILGGLIMFCVCHEDRDRRSLATVREYLTLP
PEKLRALLELMQDSDEAGGLIARAANRFLGKADREAASVLSNAQRHTHFLDSPRIAKCLARSDFAFSDLR
HRITSVFLVLPPNRMDAYSRWLRLLVSQALQDIARDAERPQNASEGRSERLTAPALFLLDEFAALGRLEA
VERAMGLMAGYGLQLWPILQDMSQLKDLYGERAGTFIANAGVQQVFGVNDFETAKWLSQMIGQETAGFQT
DSFKPGDGPSFSNNLTGRDLLTPDEIMQLPPKRQLLRVQGQATAVAQKLRYYADPEFAGLFTPETP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 5243 | GenBank | NZ_CP010775 |
| Plasmid name | pP13_h | Incompatibility group | - |
| Plasmid size | 36658 bp | Coordinate of oriT [Strand] | 33816..33835 [+] |
| Host baterium | Phaeobacter piscinae strain P13 |
Cargo genes
| Drug resistance gene | aph(6)-Id, aph(3'')-Ib |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |