Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104716 |
| Name | oriT_pL21533-4 |
| Organism | Pediococcus damnosus strain TMW 2.1533 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP012279 (5791..5811 [-], 21 nt) |
| oriT length | 21 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 21 nt
>oriT_pL21533-4
AGTGGGGTGTAAGTGCGCATT
AGTGGGGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file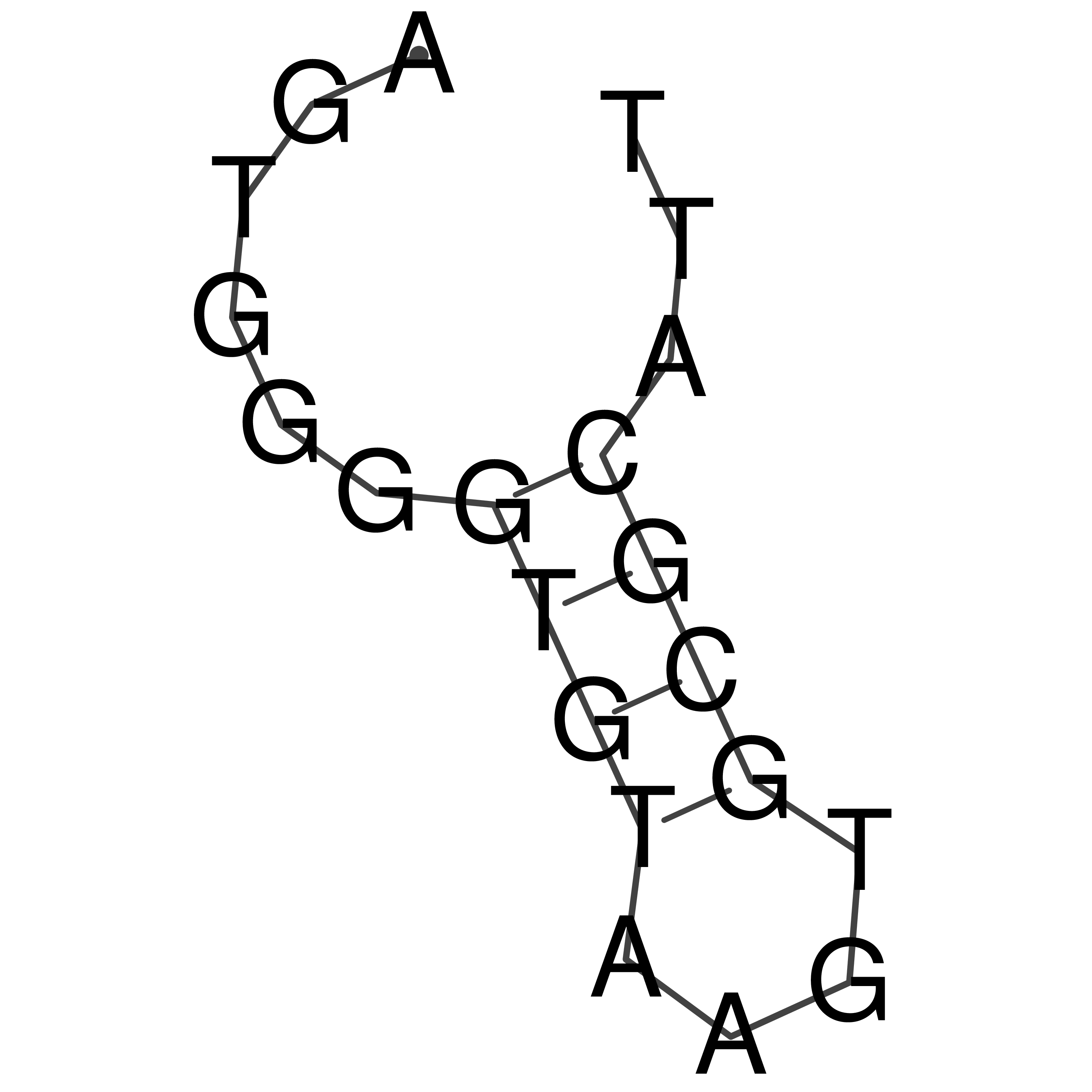
Relaxase
| ID | 3372 | GenBank | WP_062904653 |
| Name | mobQ_ADU70_RS10870_pL21533-4 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80391.90 Da Isoelectric Point: 9.1681
>WP_062904653.1 MobQ family relaxase [Pediococcus damnosus]
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYARSIMPESFILTPKNSPEWASDREQLWN
EVEKKDRKSNSRYAKEFNVALPVELSESEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNSDGTWGIKSKKQYILDDDGNKTYTGTSKYPKSRKILMVDWDKKEKITEWRHNWAASVNQALEQKNIP
DRISEKSFMEQGIDDIPTQHEGINSKRHERKAFNQQVKNYRKSKAGYKNMQEKVVNQGHLDSLSKHFSFN
EKRVVKELSHELKTYISLENLDDKRRMLFNWKNSTLIKHAIGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTQQLLAFTKRPYVGFKLL
MQQEKEVKTDLKYTLMIHSDSLESLEHVDQGLLEKYSPAEQQKITRAVKDLRTIMAVKQVIQTQYHEVLK
RAFPNGDLDDLSLVRQEQAYTAVMYYDPALKPCKVETMEQWQENPPKVFSTQEHQLGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQIFLGECKADPTIKTSQIEKIQKQLKEQQDKDDQHRKANMGHYQPLNYKPVSPSY
YLKTAFNDAIMAALYARDEDYKRQKQERGLKDTEWEMTKKKRQHQTRNRHEDGGMHL
MAIFHMSFSNISAGKGRSAIASAAYRSGEKLFDDKEGRHYFYARSIMPESFILTPKNSPEWASDREQLWN
EVEKKDRKSNSRYAKEFNVALPVELSESEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNSDGTWGIKSKKQYILDDDGNKTYTGTSKYPKSRKILMVDWDKKEKITEWRHNWAASVNQALEQKNIP
DRISEKSFMEQGIDDIPTQHEGINSKRHERKAFNQQVKNYRKSKAGYKNMQEKVVNQGHLDSLSKHFSFN
EKRVVKELSHELKTYISLENLDDKRRMLFNWKNSTLIKHAIGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTQQLLAFTKRPYVGFKLL
MQQEKEVKTDLKYTLMIHSDSLESLEHVDQGLLEKYSPAEQQKITRAVKDLRTIMAVKQVIQTQYHEVLK
RAFPNGDLDDLSLVRQEQAYTAVMYYDPALKPCKVETMEQWQENPPKVFSTQEHQLGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQIFLGECKADPTIKTSQIEKIQKQLKEQQDKDDQHRKANMGHYQPLNYKPVSPSY
YLKTAFNDAIMAALYARDEDYKRQKQERGLKDTEWEMTKKKRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3309 | GenBank | WP_016511111 |
| Name | t4cp2_ADU70_RS10930_pL21533-4 |
UniProt ID | _ |
| Length | 517 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 517 a.a. Molecular weight: 58240.39 Da Isoelectric Point: 4.9338
>WP_016511111.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactobacillaceae]
MNGTILGIVDKRIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRPPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNEYQGQPEASQETNFHQRVEKWKAKLKETAQKKAANKMSQTEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDDNNLFG
MNGTILGIVDKRIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRPPEQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMSDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDKEVADFTSFSDFDLKEIGNAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNEYQGQPEASQETNFHQRVEKWKAKLKETAQKKAANKMSQTEEEQQQDNL
DSDLERVSNKDSQTEEPEEDDDNNLFG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9372..19231
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ADU70_RS10850 (ADU70_0043) | 4504..4785 | + | 282 | WP_010620893 | type II toxin-antitoxin system RelB/DinJ family antitoxin | - |
| ADU70_RS10855 (ADU70_0044) | 4832..5017 | + | 186 | WP_010620892 | hypothetical protein | - |
| ADU70_RS11685 | 5021..5143 | + | 123 | Protein_7 | ATPase | - |
| ADU70_RS10860 (ADU70_0045) | 5133..5411 | - | 279 | WP_057707345 | hypothetical protein | - |
| ADU70_RS10865 (ADU70_0046) | 5434..5661 | - | 228 | WP_003555660 | hypothetical protein | - |
| ADU70_RS10870 (ADU70_0047) | 5911..7974 | + | 2064 | WP_062904653 | MobQ family relaxase | - |
| ADU70_RS10875 | 8057..8386 | + | 330 | WP_014216292 | hypothetical protein | - |
| ADU70_RS10880 (ADU70_0048) | 8400..9014 | + | 615 | WP_021730099 | hypothetical protein | - |
| ADU70_RS10885 (ADU70_0049) | 9016..9351 | + | 336 | WP_062904654 | CagC family type IV secretion system protein | - |
| ADU70_RS10890 (ADU70_0050) | 9372..9734 | + | 363 | WP_035457693 | hypothetical protein | trsC |
| ADU70_RS10895 (ADU70_0051) | 9703..10362 | + | 660 | WP_021730097 | TrsD/TraD family conjugative transfer protein | trsD |
| ADU70_RS10900 (ADU70_0052) | 10374..12389 | + | 2016 | WP_062904655 | DUF87 domain-containing protein | virb4 |
| ADU70_RS10905 (ADU70_0053) | 12382..13800 | + | 1419 | WP_062904656 | conjugal transfer protein | trsF |
| ADU70_RS10910 (ADU70_0054) | 13801..14955 | + | 1155 | WP_062904657 | phage tail tip lysozyme | prgK |
| ADU70_RS10915 (ADU70_0055) | 14969..15586 | + | 618 | WP_062904658 | hypothetical protein | - |
| ADU70_RS10920 (ADU70_0056) | 15573..15941 | + | 369 | WP_016511113 | hypothetical protein | - |
| ADU70_RS10925 (ADU70_0057) | 15944..16414 | + | 471 | WP_062904659 | hypothetical protein | trsJ |
| ADU70_RS10930 (ADU70_0058) | 16416..17969 | + | 1554 | WP_016511111 | VirD4-like conjugal transfer protein, CD1115 family | - |
| ADU70_RS10935 (ADU70_0059) | 17984..18373 | + | 390 | WP_035168812 | hypothetical protein | - |
| ADU70_RS10940 (ADU70_0060) | 18392..19231 | + | 840 | WP_062904660 | conjugal transfer protein TrbL family protein | trsL |
| ADU70_RS10945 (ADU70_0061) | 19246..19656 | + | 411 | WP_062904661 | hypothetical protein | - |
| ADU70_RS10950 (ADU70_0062) | 19663..21798 | + | 2136 | WP_062904662 | type IA DNA topoisomerase | - |
| ADU70_RS10955 (ADU70_0063) | 21920..22135 | + | 216 | WP_008855344 | hypothetical protein | - |
| ADU70_RS10960 (ADU70_0064) | 22139..23263 | + | 1125 | WP_062904663 | ArdC-like ssDNA-binding domain-containing protein | - |
Host bacterium
| ID | 5155 | GenBank | NZ_CP012279 |
| Plasmid name | pL21533-4 | Incompatibility group | - |
| Plasmid size | 38024 bp | Coordinate of oriT [Strand] | 5791..5811 [-] |
| Host baterium | Pediococcus damnosus strain TMW 2.1533 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |