Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104665 |
| Name | oriT_pnunamed1 |
| Organism | Latilactobacillus curvatus strain ZJUNIT8 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP029967 (55073..55092 [+], 20 nt) |
| oriT length | 20 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 20 nt
>oriT_pnunamed1
GTTGTGTGTAAGTGCGCATT
GTTGTGTGTAAGTGCGCATT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file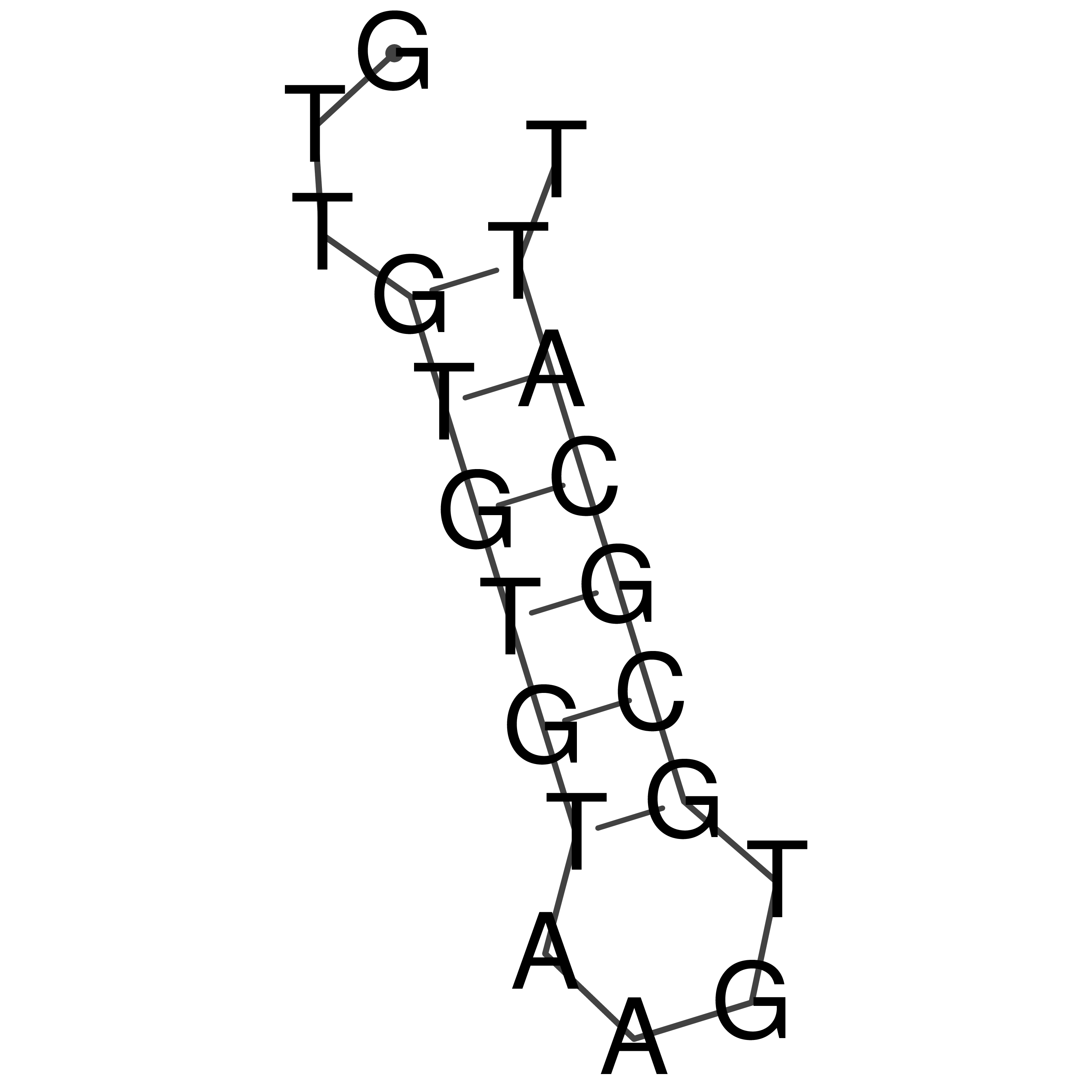
Relaxase
| ID | 3340 | GenBank | WP_111447984 |
| Name | mobQ_C0W45_RS10095_pnunamed1 |
UniProt ID | _ |
| Length | 687 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 687 a.a. Molecular weight: 80305.66 Da Isoelectric Point: 9.2830
>WP_111447984.1 MobQ family relaxase [Latilactobacillus curvatus]
MAIFHMSFQNISAGKGRSAIAGASYRSGEKLFDQKEGRSYFYARSVMPESFILTPNNAPEWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKITEWRHNWAASVNQVLEQKNIP
DRISEKSFIEQGIDDTPTQHEGINSKRHERKEFNQQVKDYRKAKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVNELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTQQLLTFTKRPYVGFKLL
MQQEKEAKIDLKYTLMIHSDSLESLEHVDKGLLEKYSPAEQQKITRAVKDLRAIMAVKQVIKTQYHEVLK
RAFPKGDLDGLPLIKQEQAYTAVMYYDPVLKPCQAETIEQWQANPPQVFSPQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKGQQAKDDQYRKANIGHYQPLNYKPVSPDY
YLKTAFSNAIMTALYARDEDYQRQKQAQGLKETEWEMTKKQRQHQTRNRHEDGGMHL
MAIFHMSFQNISAGKGRSAIAGASYRSGEKLFDQKEGRSYFYARSVMPESFILTPNNAPEWASDREKLWN
EVERKDRRANSRYAKEFNVALPVELSEDEQKELLTKYVQENFVDEGMVADVAIHRDHPDNPHAHVMLTNR
PFNPDGTWGLKSKRENILDENGNKTYTGNSRFPRSRKVWLVDWDKKEKITEWRHNWAASVNQVLEQKNIP
DRISEKSFIEQGIDDTPTQHEGINSKRHERKEFNQQVKDYRKAKASYKNNQEKVINRGHLDSLSKHFSFN
EKRVVNELSHELKTYISLESLDDKRRMLFNWKNSTLIKHAVGEDVTKQLLTINQQESSLKKADELLNKVV
DRTTKKLYPELNFEQTTQAERRELIKETDSEQTVFKGSELNERLMNIRDDLLTQQLLTFTKRPYVGFKLL
MQQEKEAKIDLKYTLMIHSDSLESLEHVDKGLLEKYSPAEQQKITRAVKDLRAIMAVKQVIKTQYHEVLK
RAFPKGDLDGLPLIKQEQAYTAVMYYDPVLKPCQAETIEQWQANPPQVFSPQEHQQGLAYLSGQLSLDQL
ENHHLQRVLKHDGTKQLFLGECKVDPTIKNSQIEKIQKQLKGQQAKDDQYRKANIGHYQPLNYKPVSPDY
YLKTAFSNAIMTALYARDEDYQRQKQAQGLKETEWEMTKKQRQHQTRNRHEDGGMHL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3281 | GenBank | WP_103127221 |
| Name | t4cp2_C0W45_RS10035_pnunamed1 |
UniProt ID | _ |
| Length | 516 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 516 a.a. Molecular weight: 57977.14 Da Isoelectric Point: 4.8681
>WP_103127221.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Lactobacillaceae]
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRAPDQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDQEVADFTSFSDFDLKDIGKAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQTEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLFG
MNGTILGIVDKQIVYQNNSTKPNRNIFVVGGPGSYKTQSVVITNLFNETENSIVVTDPKGELYEKTAGIK
LAQGYQVHVVNFANMAHSDRYNPFDYIQRDIQAETVATKIVQSENAEGKKDVWFSTQRQLLKALILFVMN
HRAPDQRNLAGVTQVLQENDVEAEEKGADSPLDTLFLDLTMTDPARRAYELGFKKAKGEMKASIIESLLA
TISKFVDQEVADFTSFSDFDLKDIGKAKVVLYVIIPVMDNTYESFINLFFSQLFDELYKLAADHHAKLPH
QVDFILDEFVNLGKFPKYEEFLATCRGYGIGVTTICQTLTQLQALYGKDKAESILGNHAVKICLNAANDV
TAKYFSDLLGKSTVKVETGSESTSHSKEESHSKSDSYSYTSRSLMTPDEIMRMPEDQSLLIFSNARPVKA
TKAFQFKLFPGADHLVNLSQNDYQGQPEASQETNFKNKVEKWKAKLKETAQKEVANKMSQTEEEQQQDNL
DSDLERVSNKDSQTEAPEEDDNNLFG
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 41650..51496
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| C0W45_RS09995 (C0W45_09995) | 36699..37142 | - | 444 | WP_016526921 | hypothetical protein | - |
| C0W45_RS10480 (C0W45_10000) | 37405..37467 | + | 63 | WP_230083882 | putative holin-like toxin | - |
| C0W45_RS10005 (C0W45_10005) | 37620..38744 | - | 1125 | WP_111447974 | ArdC-like ssDNA-binding domain-containing protein | - |
| C0W45_RS10010 (C0W45_10010) | 38747..38962 | - | 216 | WP_111447975 | hypothetical protein | - |
| C0W45_RS10015 (C0W45_10015) | 39082..41217 | - | 2136 | WP_111447976 | type IA DNA topoisomerase | - |
| C0W45_RS10020 (C0W45_10020) | 41224..41634 | - | 411 | WP_111447977 | hypothetical protein | - |
| C0W45_RS10025 (C0W45_10025) | 41650..42489 | - | 840 | WP_162619244 | conjugal transfer protein TrbL family protein | trsL |
| C0W45_RS10030 (C0W45_10030) | 42513..42887 | - | 375 | WP_025085237 | hypothetical protein | - |
| C0W45_RS10035 (C0W45_10035) | 42902..44452 | - | 1551 | WP_103127221 | VirD4-like conjugal transfer protein, CD1115 family | - |
| C0W45_RS10040 (C0W45_10040) | 44454..44924 | - | 471 | WP_111447979 | conjugal transfer protein | trsJ |
| C0W45_RS10045 (C0W45_10045) | 44927..45295 | - | 369 | WP_111447980 | thioredoxin family protein | - |
| C0W45_RS10050 (C0W45_10050) | 45282..45899 | - | 618 | WP_103127218 | hypothetical protein | - |
| C0W45_RS10055 (C0W45_10055) | 45913..47067 | - | 1155 | WP_111447981 | phage tail tip lysozyme | orf14 |
| C0W45_RS10060 (C0W45_10060) | 47068..48486 | - | 1419 | WP_111447982 | type VII secretion protein EssB/YukC | trsF |
| C0W45_RS10065 (C0W45_10065) | 48479..50494 | - | 2016 | WP_111447983 | DUF87 domain-containing protein | virb4 |
| C0W45_RS10070 (C0W45_10070) | 50506..51165 | - | 660 | WP_025085233 | TrsD/TraD family conjugative transfer protein | trsD |
| C0W45_RS10075 (C0W45_10075) | 51134..51496 | - | 363 | WP_014216295 | hypothetical protein | trsC |
| C0W45_RS10080 (C0W45_10080) | 51518..51853 | - | 336 | WP_076648311 | CagC family type IV secretion system protein | - |
| C0W45_RS10085 (C0W45_10085) | 51855..52469 | - | 615 | WP_039098895 | hypothetical protein | - |
| C0W45_RS10485 (C0W45_10090) | 52511..52822 | - | 312 | WP_039098894 | hypothetical protein | - |
| C0W45_RS10095 (C0W45_10095) | 52907..54970 | - | 2064 | WP_111447984 | MobQ family relaxase | - |
Host bacterium
| ID | 5104 | GenBank | NZ_CP029967 |
| Plasmid name | pnunamed1 | Incompatibility group | - |
| Plasmid size | 55352 bp | Coordinate of oriT [Strand] | 55073..55092 [+] |
| Host baterium | Latilactobacillus curvatus strain ZJUNIT8 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21 |