Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104642 |
| Name | oriT_FDAARGOS_1271|unnamed |
| Organism | Salmonella enterica strain FDAARGOS_1271 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP069519 (84600..84750 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 12..17, 24..29 (AACCCG..CGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_FDAARGOS_1271|unnamed
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
TGGTCGATGTGAACCCGTTTGACCGGGTTATGAATGAGTTGAAAAGTCGTGGCCGCAAGAACGCTCATATCCTGAGCATCCTTCAATTCGACTGGCCCGCATCGGAGGTCATCATCGAGAAGCTGAGCTGTTACATCACAGATGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file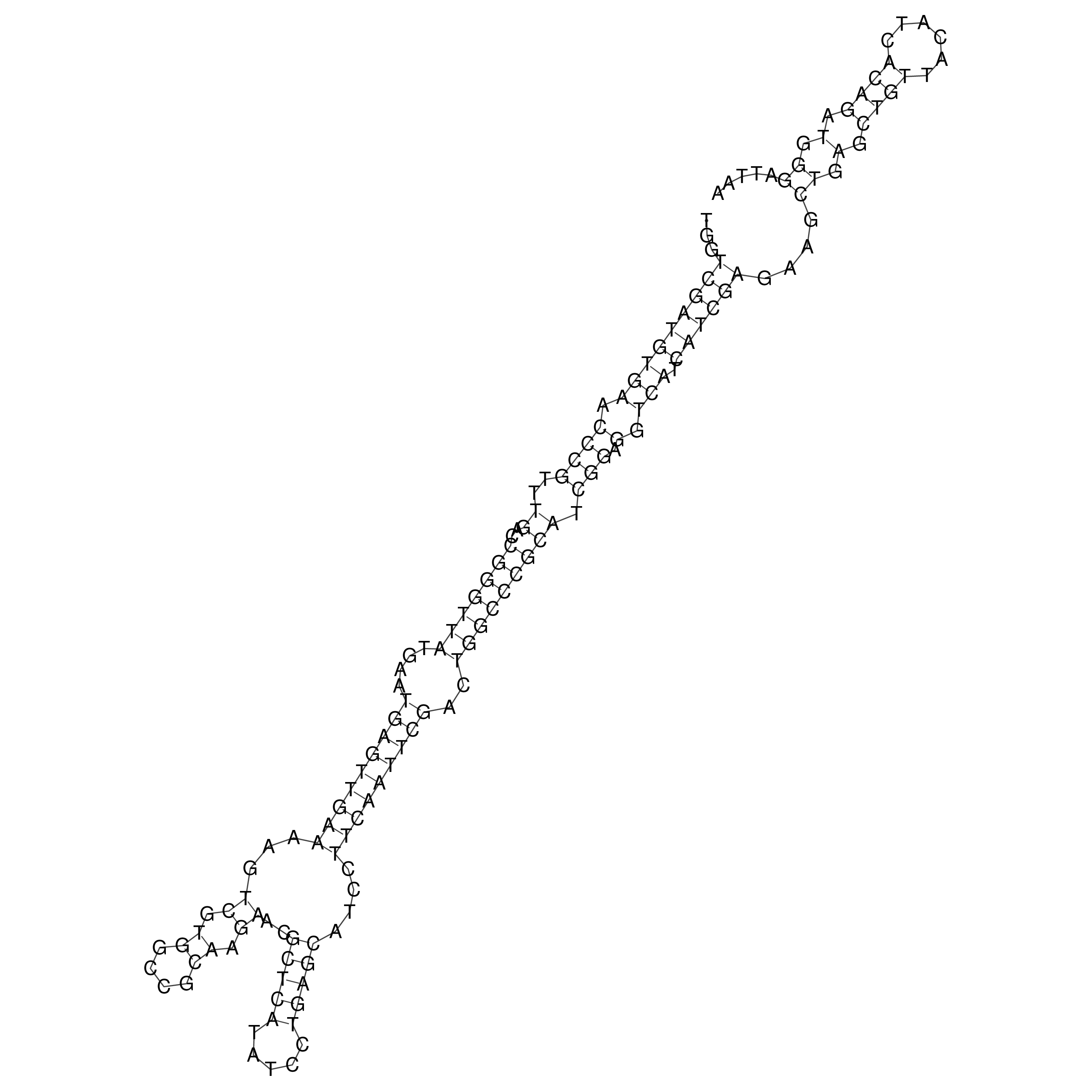
Relaxase
| ID | 3333 | GenBank | WP_000909947 |
| Name | mobH_I6K01_RS23115_FDAARGOS_1271|unnamed |
UniProt ID | _ |
| Length | 987 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 987 a.a. Molecular weight: 109404.96 Da Isoelectric Point: 4.8922
>WP_000909947.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLEIPDITEPEKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVAPKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
MLKALHKLFGGRSGAIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWATQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAYRHGIDRYFIRWRDKRHKRHEQFSL
LAWHRIIPAETQEFLSKSGPSIIEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVNEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISQDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGPVKILMLRLESNDLVFTTEPPAAVTGEVVGDVEDAEIE
FVDPEEADEDQAEGDAGLNNDMLAAELEAEKALAGLGFGDAMAMLKSTSDIAEEKPEQEDTESTELPKPD
ASKKGKQQSKPGKARAEKQPQKPEVKEDLSPQDIAKNAPPLANDDPLQALKDVGGGLGDIDFPFDVFNAS
AETACTDATNLEIPDITEPEKQEEQPKPDFVPQEQNSLQNGDFPIFSDSDEPPSWAIEPLPMLTDTPEQT
IPAPDMQHAGKPKELEKDAKTLLSEMLAGYGEASTLLEQAIMPVLEGKTTLGEVLCLMKGQAVILYPEGA
RSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLVDQLSDAVVAAIKDAEASMGGYQEAFELV
SPPGSGVSKNKSAPKQQSRKKTQQQKPEVDAGAGSEQKTKDKSLQQQPKEKQGNVASLVEEQKRKPVQDQ
EKQNVARLPKREAQPVAPKPKVEHERELGHVEVRERDEPEVREFEPPKAKTNPKDINEEDFLPSGVTPEK
ALLMLKDMIQKRSGRWLVTPVLEEDGCLVTSVKAFDMIAGENVGISKHILYGLLSRAQRRPLLKKRQGKL
YLEVDKA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3273 | GenBank | WP_000178858 |
| Name | traD_I6K01_RS23120_FDAARGOS_1271|unnamed |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69248.76 Da Isoelectric Point: 7.1093
>WP_000178858.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWMAASCAALAVEQVSTMPSEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPEDMWLGSGFLWENRHAQRVFEILKRDWTSIVGKESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVLTHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVKPDWEAEAAAYLEKAKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNTDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3274 | GenBank | WP_000587426 |
| Name | traC_I6K01_RS23200_FDAARGOS_1271|unnamed |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92489.76 Da Isoelectric Point: 5.9204
>WP_000587426.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRILAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQHLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
MIITIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDQSVGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTSLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRILAKTSKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQHLAQIRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYDDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSQKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSTFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSDGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRKQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 82684..107788
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| I6K01_RS23105 (I6K01_23105) | 78724..78957 | - | 234 | WP_001191893 | hypothetical protein | - |
| I6K01_RS23110 (I6K01_23110) | 78939..79556 | - | 618 | WP_079778879 | hypothetical protein | - |
| I6K01_RS23115 (I6K01_23115) | 79724..82687 | + | 2964 | WP_000909947 | MobH family relaxase | - |
| I6K01_RS23120 (I6K01_23120) | 82684..84549 | + | 1866 | WP_000178858 | conjugative transfer system coupling protein TraD | virb4 |
| I6K01_RS23125 (I6K01_23125) | 84599..85144 | + | 546 | WP_023356350 | hypothetical protein | - |
| I6K01_RS23130 (I6K01_23130) | 85101..85730 | + | 630 | WP_000743448 | DUF4400 domain-containing protein | tfc7 |
| I6K01_RS23135 (I6K01_23135) | 85741..86187 | + | 447 | WP_000122583 | hypothetical protein | - |
| I6K01_RS23140 (I6K01_23140) | 86197..86574 | + | 378 | WP_000869260 | hypothetical protein | - |
| I6K01_RS23145 (I6K01_23145) | 86574..87236 | + | 663 | WP_000045163 | hypothetical protein | - |
| I6K01_RS23150 (I6K01_23150) | 87415..87558 | + | 144 | WP_023356351 | hypothetical protein | - |
| I6K01_RS23155 (I6K01_23155) | 87571..87918 | + | 348 | WP_001102160 | hypothetical protein | - |
| I6K01_RS23160 (I6K01_23160) | 88061..88342 | + | 282 | WP_000805626 | type IV conjugative transfer system protein TraL | traL |
| I6K01_RS23165 (I6K01_23165) | 88339..88965 | + | 627 | WP_001049718 | TraE/TraK family type IV conjugative transfer system protein | traE |
| I6K01_RS23170 (I6K01_23170) | 88949..89866 | + | 918 | WP_000839951 | type-F conjugative transfer system secretin TraK | traK |
| I6K01_RS23175 (I6K01_23175) | 89863..91179 | + | 1317 | WP_000595204 | TraB/VirB10 family protein | traB |
| I6K01_RS23180 (I6K01_23180) | 91176..91754 | + | 579 | WP_000794193 | type IV conjugative transfer system lipoprotein TraV | traV |
| I6K01_RS23185 (I6K01_23185) | 91758..92150 | + | 393 | WP_012766327 | TraA family conjugative transfer protein | - |
| I6K01_RS23190 (I6K01_23190) | 92351..97837 | + | 5487 | WP_000606836 | Ig-like domain-containing protein | - |
| I6K01_RS23195 (I6K01_23195) | 97986..98693 | + | 708 | WP_001259344 | DsbC family protein | - |
| I6K01_RS23200 (I6K01_23200) | 98690..101137 | + | 2448 | WP_000587426 | type IV secretion system protein TraC | virb4 |
| I6K01_RS23205 (I6K01_23205) | 101152..101469 | + | 318 | WP_000364398 | hypothetical protein | - |
| I6K01_RS23210 (I6K01_23210) | 101466..101996 | + | 531 | WP_000672456 | S26 family signal peptidase | - |
| I6K01_RS23215 (I6K01_23215) | 101959..103227 | + | 1269 | WP_001553264 | TrbC family F-type conjugative pilus assembly protein | traW |
| I6K01_RS23220 (I6K01_23220) | 103224..103901 | + | 678 | WP_000577801 | EAL domain-containing protein | - |
| I6K01_RS23225 (I6K01_23225) | 103895..104899 | + | 1005 | WP_000911304 | TraU family protein | traU |
| I6K01_RS23230 (I6K01_23230) | 105050..107788 | + | 2739 | WP_001256493 | conjugal transfer mating pair stabilization protein TraN | traN |
| I6K01_RS23235 (I6K01_23235) | 107827..108687 | - | 861 | WP_000679674 | hypothetical protein | - |
| I6K01_RS23240 (I6K01_23240) | 108799..109449 | - | 651 | WP_000796641 | hypothetical protein | - |
| I6K01_RS23245 (I6K01_23245) | 109740..110060 | + | 321 | WP_000547567 | hypothetical protein | - |
| I6K01_RS23250 (I6K01_23250) | 110371..110556 | + | 186 | WP_001186892 | hypothetical protein | - |
| I6K01_RS23255 (I6K01_23255) | 110777..111745 | + | 969 | WP_001553257 | AAA family ATPase | - |
| I6K01_RS23260 (I6K01_23260) | 111757..112662 | + | 906 | WP_000739141 | hypothetical protein | - |
Host bacterium
| ID | 5081 | GenBank | NZ_CP069519 |
| Plasmid name | FDAARGOS_1271|unnamed | Incompatibility group | IncA/C |
| Plasmid size | 128163 bp | Coordinate of oriT [Strand] | 84600..84750 [+] |
| Host baterium | Salmonella enterica strain FDAARGOS_1271 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |