Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104620 |
| Name | oriT_pG32-51 |
| Organism | Proteus terrae subsp. cibarius strain G32 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP053373 (44280..44320 [+], 41 nt) |
| oriT length | 41 nt |
| IRs (inverted repeats) | 14..22, 29..37 (TAAAGTATA..TATACTTTA) 2..8, 13..19 (ACTTTAT..ATAAAGT) |
| Location of nic site | 27..28 |
| Conserved sequence flanking the nic site |
GTGTGTTATA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 41 nt
>oriT_pG32-51
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
CACTTTATGAATATAAAGTATAGTGTGTTATACTTTACATG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file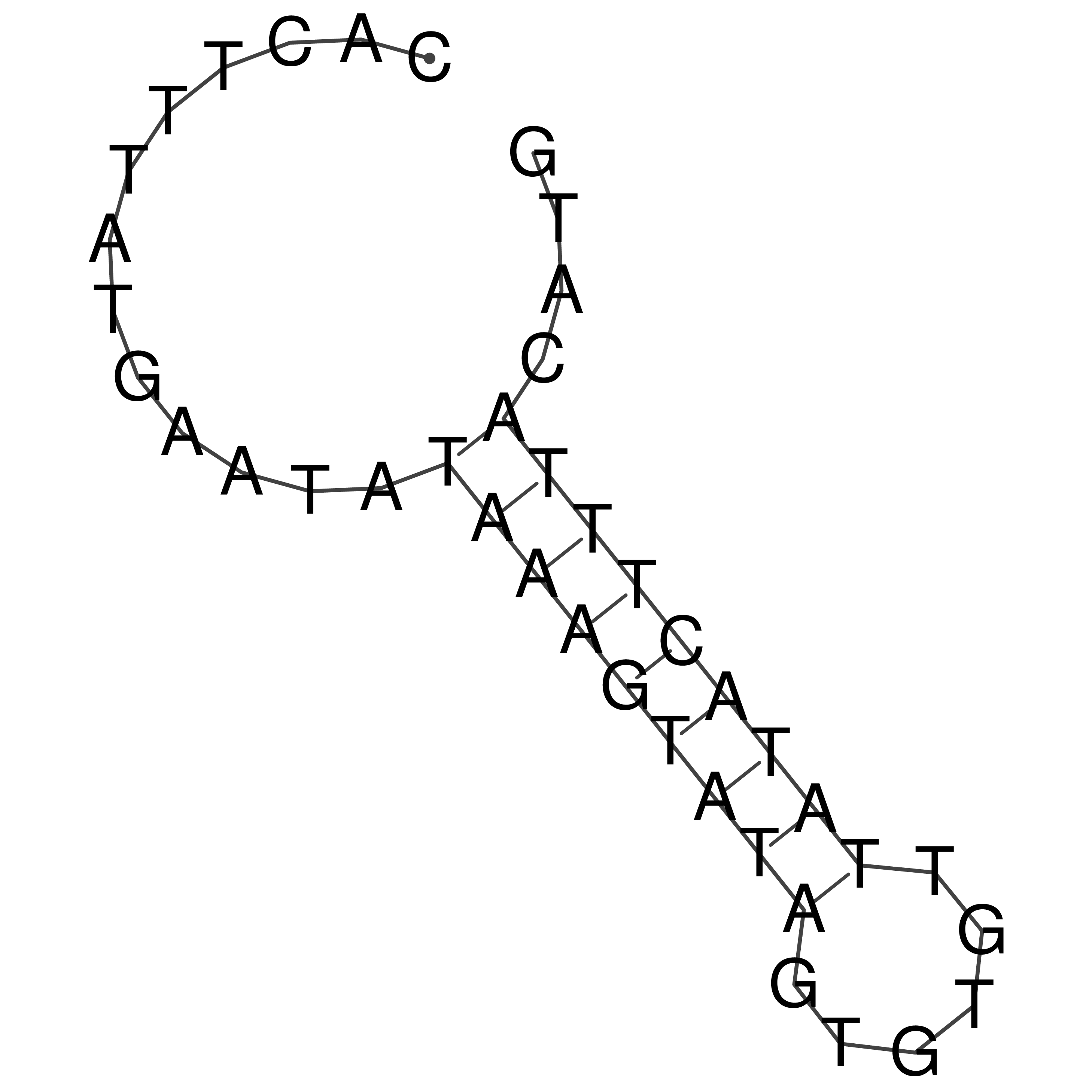
Relaxase
| ID | 3320 | GenBank | WP_012779614 |
| Name | mobV_HND96_RS19775_pG32-51 |
UniProt ID | C7C2U1 |
| Length | 408 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 408 a.a. Molecular weight: 48229.55 Da Isoelectric Point: 7.0097
>WP_012779614.1 MULTISPECIES: MobV family relaxase [Bacteria]
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKDQIDKNDKDIDDYNDLVKRYNNLYEENTSLKSQIGDLKEEIKLIY
QSTKRFLKDRISDFKAFKEVFKELADNISNISREKGLDSSFKKEFDRENKKKQTRGIR
MSMIVARMQKMKAENLVGIGNHNQRKTKNHSNPDIDTSLSKLNYDLVDRTQNYKTDIENFINENKSTTRA
VRKDAVLVNEWIISSDKDFFDNLTESEIENFFERSKDYFAEKFGEKNIRYATVHLDESTPHMHMGIVPFD
KDNKLSAKRVFNRQALRDVQEELPKYLQDFQFEIARGQKGSERKNLTVPEFKKLKEEELEIKKELQIKKD
ELIAYTKENQIDKKLDITPIKEMEDIEIETDEKTLFGKNKTEIVRQWTGNIILSENDYLKLNKEIKKGKK
TEGRLAAILETDVYQENKELKNELKDQIDKNDKDIDDYNDLVKRYNNLYEENTSLKSQIGDLKEEIKLIY
QSTKRFLKDRISDFKAFKEVFKELADNISNISREKGLDSSFKKEFDRENKKKQTRGIR
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | C7C2U1 |
T4CP
| ID | 3258 | GenBank | WP_159364216 |
| Name | t4cp2_HND96_RS19510_pG32-51 |
UniProt ID | _ |
| Length | 670 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 670 a.a. Molecular weight: 76242.39 Da Isoelectric Point: 8.9435
>WP_159364216.1 type IV secretory system conjugative DNA transfer family protein [Proteus terrae]
MLNKKKIAIFLILLFLISVGASFYTSSMALLKMVGMPISFETVKWNTFIDTYKLYAGQKAYQKYLLAGMA
GGIAPFVLYLGFIILIIIGLKPKKTLHGDARLATDMDLAKSGFFPTKNKPNKKASTAILIGKMDKGRYKG
QYIQYMGQQFLMLYAPTRSGKGVGIVIPNCLYYFESLVVLDIKLENFISTAGHRKDKLNQEVFLFCPDGY
LDSEDMKDKKLRSHRYNPLFYIRRDPINRFGDLTKISNILFPLTGDKNDMWTDLSANVFNALVLFLLDTE
SETVIEDIEIEKDGVVHYEERIKPKYKVTMSQVFGLSVPKDGTTLGDWFINEIATRNTKENKDAWESYIA
SEKSGNKSVTRPELCLLSDDTVTLMRQFSQQKPEQQQSIMLTFNANMKMFANPVTAAATDGNDFDLRDVR
RKRMTVYFGLAPAALSQYARLTNLFFSQLLNENVRTLPEHDSSLKYQCLMMLDEFTSMGCLDIIQVSLAF
TAGYNMRFVFILQNQEQLFDEKKGYGKNGGNTILKNCAVELFYPPKEVDESVKKMSETLGYKDMKVISTS
VSKGKSASKSTSTSIQKRALMLPQEIIELRDMKHSSGMALREIVKSEFTRPFVANKIVYFAEPYFSERKK
YAETHIPDIPVLNIEQDELAKALHEVILRDANHIVEPDFD
MLNKKKIAIFLILLFLISVGASFYTSSMALLKMVGMPISFETVKWNTFIDTYKLYAGQKAYQKYLLAGMA
GGIAPFVLYLGFIILIIIGLKPKKTLHGDARLATDMDLAKSGFFPTKNKPNKKASTAILIGKMDKGRYKG
QYIQYMGQQFLMLYAPTRSGKGVGIVIPNCLYYFESLVVLDIKLENFISTAGHRKDKLNQEVFLFCPDGY
LDSEDMKDKKLRSHRYNPLFYIRRDPINRFGDLTKISNILFPLTGDKNDMWTDLSANVFNALVLFLLDTE
SETVIEDIEIEKDGVVHYEERIKPKYKVTMSQVFGLSVPKDGTTLGDWFINEIATRNTKENKDAWESYIA
SEKSGNKSVTRPELCLLSDDTVTLMRQFSQQKPEQQQSIMLTFNANMKMFANPVTAAATDGNDFDLRDVR
RKRMTVYFGLAPAALSQYARLTNLFFSQLLNENVRTLPEHDSSLKYQCLMMLDEFTSMGCLDIIQVSLAF
TAGYNMRFVFILQNQEQLFDEKKGYGKNGGNTILKNCAVELFYPPKEVDESVKKMSETLGYKDMKVISTS
VSKGKSASKSTSTSIQKRALMLPQEIIELRDMKHSSGMALREIVKSEFTRPFVANKIVYFAEPYFSERKK
YAETHIPDIPVLNIEQDELAKALHEVILRDANHIVEPDFD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 4724..14234
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HND96_RS19500 (HND96_19555) | 1727..2059 | - | 333 | WP_050878446 | hypothetical protein | - |
| HND96_RS19505 (HND96_19560) | 2123..2656 | - | 534 | WP_050878443 | hypothetical protein | - |
| HND96_RS19510 (HND96_19565) | 2666..4678 | - | 2013 | WP_159364216 | type IV secretory system conjugative DNA transfer family protein | - |
| HND96_RS19515 (HND96_19570) | 4724..5755 | - | 1032 | WP_050878441 | P-type DNA transfer ATPase VirB11 | virB11 |
| HND96_RS19520 (HND96_19575) | 5804..7015 | - | 1212 | WP_238795221 | TrbI/VirB10 family protein | virB10 |
| HND96_RS19525 (HND96_19580) | 7015..7809 | - | 795 | WP_050878438 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| HND96_RS19530 (HND96_19585) | 7809..8567 | - | 759 | WP_050878436 | type IV secretion system protein | virB8 |
| HND96_RS19535 (HND96_19590) | 8574..8708 | - | 135 | WP_156734538 | lipoprotein | - |
| HND96_RS19540 (HND96_19595) | 8729..9733 | - | 1005 | WP_050878434 | type IV secretion system protein | virB6 |
| HND96_RS19545 (HND96_19600) | 9783..10472 | - | 690 | WP_050878432 | type IV secretion system protein | - |
| HND96_RS19550 (HND96_19605) | 10469..12907 | - | 2439 | WP_072100189 | VirB4 family type IV secretion system protein | virb4 |
| HND96_RS19555 (HND96_19610) | 12911..13225 | - | 315 | WP_050878430 | VirB3 family type IV secretion system protein | virB3 |
| HND96_RS19560 (HND96_19615) | 13274..13612 | - | 339 | WP_229764494 | TrbC/VirB2 family protein | virB2 |
| HND96_RS19565 (HND96_19620) | 13614..14234 | - | 621 | WP_080366081 | lytic transglycosylase domain-containing protein | virB1 |
| HND96_RS19570 (HND96_19625) | 14393..15922 | - | 1530 | WP_156734535 | YadA-like family protein | - |
| HND96_RS19575 (HND96_19630) | 15994..16317 | - | 324 | WP_195831643 | hypothetical protein | - |
| HND96_RS19580 (HND96_19635) | 16495..16788 | - | 294 | WP_050878422 | sigma factor-like helix-turn-helix DNA-binding protein | - |
| HND96_RS19585 (HND96_19640) | 16896..17231 | - | 336 | WP_050878420 | hypothetical protein | - |
| HND96_RS19590 (HND96_19645) | 17328..18155 | - | 828 | WP_050878418 | GIY-YIG nuclease family protein | - |
| HND96_RS19595 (HND96_19650) | 18165..18995 | - | 831 | WP_050878417 | replication initiation protein | - |
Host bacterium
| ID | 5059 | GenBank | NZ_CP053373 |
| Plasmid name | pG32-51 | Incompatibility group | - |
| Plasmid size | 51686 bp | Coordinate of oriT [Strand] | 44280..44320 [+] |
| Host baterium | Proteus terrae subsp. cibarius strain G32 |
Cargo genes
| Drug resistance gene | cfr |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |