Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104614 |
| Name | oriT_pVCR94 |
| Organism | Vibrio cholerae strain E4 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP033514 (16873..17023 [+], 151 nt) |
| oriT length | 151 nt |
| IRs (inverted repeats) | 93..98, 106..111 (TGGCCT..AGGCCA) 12..17, 24..29 (AACCCT..AGGGTT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 151 nt
>oriT_pVCR94
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
TGGTCGATGTGAACCCTTTCGACAGGGTTATGAATGAATTGAAAAGTCGTGGCCGCAAGAACGCTCACATCCTGAGCATCCTCCAATTCGACTGGCCTGCATCGGAGGCCATCATCGAGAAGCTGAGCTGCTACATCACAGACGGGATTAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file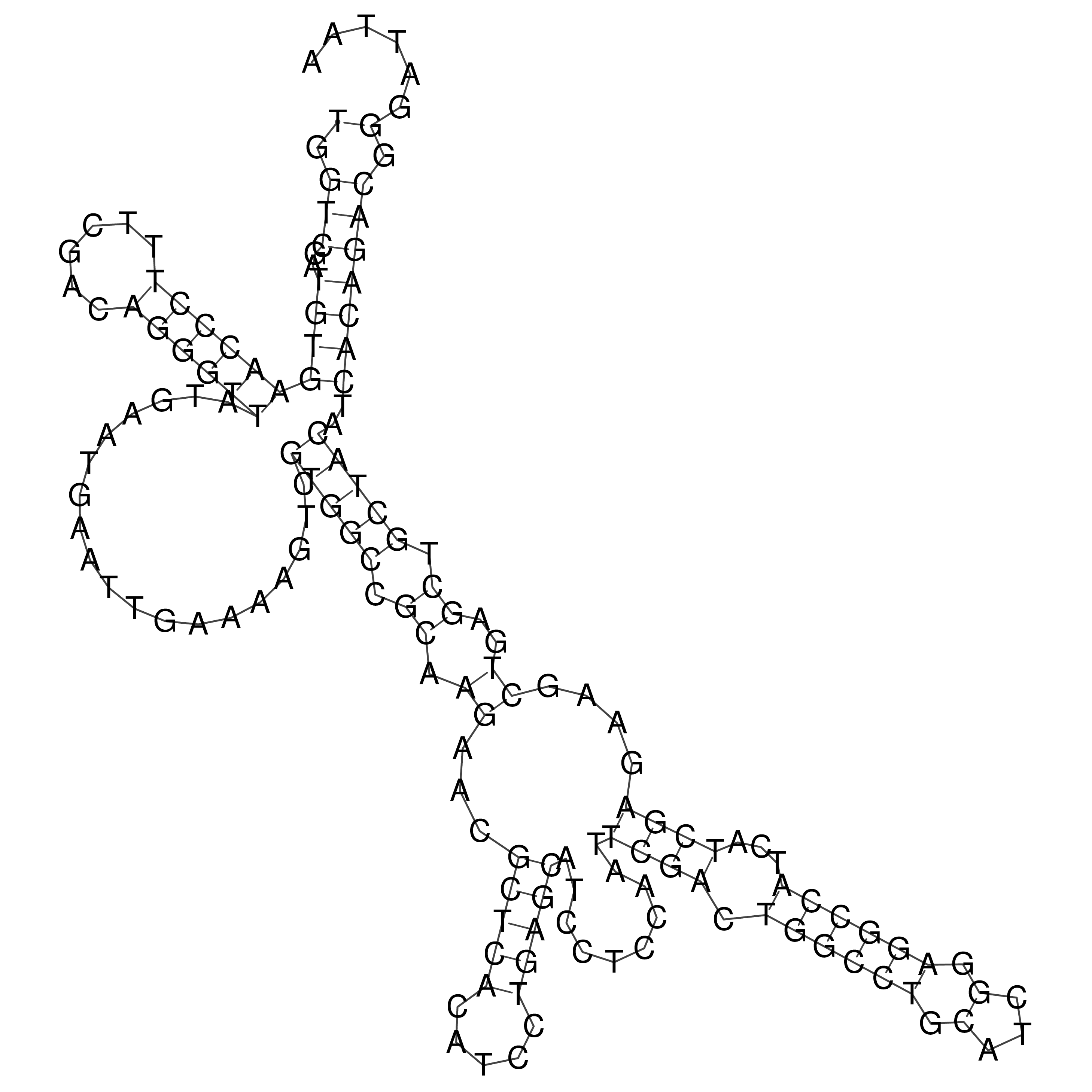
Relaxase
| ID | 3316 | GenBank | WP_011872892 |
| Name | mobH_EEL44_RS05400_pVCR94 |
UniProt ID | A0A1W6AQM7 |
| Length | 992 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 992 a.a. Molecular weight: 109595.88 Da Isoelectric Point: 4.8967
>WP_011872892.1 MULTISPECIES: MobH family relaxase [Gammaproteobacteria]
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
MLKALNKLFGGRSGVIETAPSARVLPLKDVEDEEIPRYPPFAKGLPVAPLDKILATQAELIEKVRNSLGF
TVDDFNRLVLPVIQRYAAFVHLLPASESHHHRGAGGLFRHGLEVAFWAAQASESVIFSIEGTPRERRDNE
PRWRLASCFSGLLHDVGKPLSDVSITDKDGSITWNPYSESLHDWAHRHEIDRYFIRWRDKRHKRHEQFSL
LAVDRIIPAETREFLSKSGPSIMEAMLEAISGTSVNQPVTKLMLRADQESVSRDLRQSRLDVDEFSYGVP
VERYVFDAIRRLVKTGKWKVNEPGAKVWHLNQGVFIAWKQLGDLYDLISHDKIPGIPRDPDTLADILIER
GFAVPNTVQEKGERAYYRYWEVLPEMLQEAAGSVKILMLRLESNDLVFTTEPPAAVAAEVVGDVEDAEIE
FVDPEEADDGDDQEEGEAALNDDMLAAEQEAEKALAGLGFGDAMEMLKSTSDAVEEKPEQKDAGPTESSK
PDAGKKGKPQSKPGKAKPKSDTEKQPHKPEAKEDLSPQDIAKNAPPLANDNPLQALKDVGGGLGDIDFPF
DAFNASAETTSTDATNSEIPDVAMPGKQEEQPKQDFVPQEQNSLQGDDFPMFGGSDEPPSWAIEPLPMLT
DAPEQPTHTPEMPHTDNVNQHEKDAKTLLVEMLSGYGEASALLEQAIMPVLEGKTTLGEVLCLMKGQAVI
LYPDGARSLGAPSEVLSKLSHANAIVPDPIMPGRKVRDFSGVKAIVLAEQLSDAVVAAIKDAEASMGGYQ
DAFELVSPPGLDASKNKSAPKQQSRKKAQQQKPEVNAGKASPEQKAKGKDSQPQPKEKKVDVTSPVEEQQ
RKPVQEKQNVARLPKREAQPVAPEPKVEREKELGHVEVREREEPEVREFEPPKAKTNPKDINAEDFLPSG
VTPQKALQMLKDMIQKRSGRWLVTPVLEEDGCLVTSDKAFDMIAGENIGISKHILCGMLSRAQRRPLLKK
RQGKLYLEVNET
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1W6AQM7 |
T4CP
| ID | 3256 | GenBank | WP_000178857 |
| Name | traD_EEL44_RS05405_pVCR94 |
UniProt ID | _ |
| Length | 621 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 621 a.a. Molecular weight: 69225.71 Da Isoelectric Point: 6.9403
>WP_000178857.1 MULTISPECIES: conjugative transfer system coupling protein TraD [Gammaproteobacteria]
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKVKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
MTMSYDPLAYEMPWRPNYEKNAVAGWLAASGAALAVEQVSTMPPEPFYWMTGICGVMAMARLPKAIKLHL
LQKHLKGRDLEFISIAELQKYIKDTPDDMWLGSGFLWENRHAQRVFEILKRDWTSIVGRESTVKKVVRKI
QGKKKELPIGQPWIHGVEPKEEKLMQPLKHTEGHSLIVGTTGSGKTRMFDILISQAILRGEAVIIIDPKG
DKEMRDNARRACEAMGQPERFVSFHPAFPEESVRIDPLRNFTRVTEIASRLAALIPSEAGADPFKSFGWQ
ALNNIAQGLVITHDRPNLTKLRRFLEGGAAGLVIKAVQAYSERVMPDWEAEAAAYLEKVKNGSREKIAFA
LMKFYYDIIQPEHPNSDLEGLLSMFQHDQTHFSKMVANLLPIMNMLTSGELGPLLSPDSSDLSDERQITD
SAKIINNAQVAYLGLDSLTDNMVGSAMGSIFLSDLTAVAGDRYNYGVNNRPVNIFVDEAAEVINDPFIQL
LNKGRGAKLRLFVATQTFADFAARLGSKDKALQVLGNINNTFALRIVDGETQEYIADNLPKTRLKYVMRT
QGQNSDGKEPIMHGGNQGERLMEEEADLFPAQLLGMLPNLEYIAKISGGTIVKGRLPILTQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3257 | GenBank | WP_011872839 |
| Name | traC_EEL44_RS05480_pVCR94 |
UniProt ID | _ |
| Length | 815 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 815 a.a. Molecular weight: 92576.85 Da Isoelectric Point: 5.9216
>WP_011872839.1 MULTISPECIES: type IV secretion system protein TraC [Gammaproteobacteria]
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
MIVTIKKKLEETLIPEHLRAAGIIPVLAYDEDDHVFLMDDHSAGFGFMCEPLCGADEKVQERMNGFLNQE
FPSKTTLQFVLFRSPDINQEMYRMMGLRDGFRHELLTSVIKERINFLQHHTTDRIFAKTNKGIYDNGLIQ
DLKLFVTCKVPIKNNNPTESELQQLAQLRTKVESSLQTVGLRPRTMTAVNYIRIMSTILNWGPDASWRHD
SVDWEMDKPICEQIFDYGTDVEVSKNGIRLGDYHAKVMSAKKLPDVFYFGDALTYAGDLSGGNSSIKENY
MVVTNVFFPEAESTKNTLERKRQFTVNQAYGPMLKFVPVLADKKESFDTLYESMKEGAKPVKITYSVVLF
APTKERVEAAAMAARNIWRESRFELMEDKFVALPMFLNCLPFCTDRDAVRDLFRYKTMTTEQAAVVLPVF
GEWKGTGTYHAALISRNGQLMSLSLHDSNTNKNLVIAAESGSGKSFLTNELIFSYLSEGAQVWVIDAGKS
YQKLSEMLNGDFVHFEEGTHVCLNPFELIQNYEDEEDAIVSLVCAMASAKGLLDEWQISALKQVLSRLWE
EKGKEMKVDDIAERCLEEENDQRLKDIGQQLYAFTSKGSYGKYFSRKNNVSFQNQFTVLELDELQGRKHL
RQVVLLQLIYQIQQEVFLGERNRKKVVIVDEAWDLLKEGEVSVFMEHAYRKFRKYGGSVVIATQSINDLY
ENAVGRAIAENSASMYLLGQTEETVESVKRSGRLTLSEGGFHTLKTVHTIQGVYSEIFIKSKSGMGVGRL
IVGDFQKLLYSTDPVDVNAIDQFVKQGMSIPEAIKAVMRSRQQAA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 14957..40147
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EEL44_RS05390 (EEL44_05390) | 10983..11216 | - | 234 | WP_001191892 | hypothetical protein | - |
| EEL44_RS05395 (EEL44_05395) | 11198..11815 | - | 618 | WP_001249396 | hypothetical protein | - |
| EEL44_RS05400 (EEL44_05400) | 11982..14960 | + | 2979 | WP_011872892 | MobH family relaxase | - |
| EEL44_RS05405 (EEL44_05405) | 14957..16822 | + | 1866 | WP_000178857 | conjugative transfer system coupling protein TraD | virb4 |
| EEL44_RS05410 (EEL44_05410) | 16872..17417 | + | 546 | WP_000228720 | hypothetical protein | - |
| EEL44_RS05415 (EEL44_05415) | 17374..18003 | + | 630 | WP_000743450 | DUF4400 domain-containing protein | tfc7 |
| EEL44_RS05420 (EEL44_05420) | 18013..18459 | + | 447 | WP_000122506 | hypothetical protein | - |
| EEL44_RS05425 (EEL44_05425) | 18469..18846 | + | 378 | WP_000869261 | hypothetical protein | - |
| EEL44_RS05430 (EEL44_05430) | 18846..19508 | + | 663 | WP_021293632 | hypothetical protein | - |
| EEL44_RS20995 | 19689..19832 | + | 144 | WP_001275801 | hypothetical protein | - |
| EEL44_RS05435 (EEL44_05435) | 19844..20209 | + | 366 | WP_001052531 | hypothetical protein | - |
| EEL44_RS05440 (EEL44_05440) | 20354..20634 | + | 281 | Protein_25 | type IV conjugative transfer system protein TraL | - |
| EEL44_RS05445 (EEL44_05445) | 20631..21257 | + | 627 | WP_001049716 | TraE/TraK family type IV conjugative transfer system protein | traE |
| EEL44_RS05450 (EEL44_05450) | 21241..22158 | + | 918 | WP_000794249 | type-F conjugative transfer system secretin TraK | traK |
| EEL44_RS05455 (EEL44_05455) | 22158..23471 | + | 1314 | WP_164491403 | TraB/VirB10 family protein | traB |
| EEL44_RS05460 (EEL44_05460) | 23468..24046 | + | 579 | WP_000793435 | type IV conjugative transfer system lipoprotein TraV | traV |
| EEL44_RS05465 (EEL44_05465) | 24050..24442 | + | 393 | WP_069369997 | TraA family conjugative transfer protein | - |
| EEL44_RS05470 (EEL44_05470) | 24643..30174 | + | 5532 | WP_024126918 | Ig-like domain-containing protein | - |
| EEL44_RS05475 (EEL44_05475) | 30323..31030 | + | 708 | WP_001259347 | DsbC family protein | - |
| EEL44_RS05480 (EEL44_05480) | 31027..33474 | + | 2448 | WP_011872839 | type IV secretion system protein TraC | virb4 |
| EEL44_RS05485 (EEL44_05485) | 33489..33806 | + | 318 | WP_011872858 | hypothetical protein | - |
| EEL44_RS05490 (EEL44_05490) | 33803..34333 | + | 531 | WP_021293639 | S26 family signal peptidase | - |
| EEL44_RS05495 (EEL44_05495) | 34296..35561 | + | 1266 | WP_121926692 | TrbC family F-type conjugative pilus assembly protein | traW |
| EEL44_RS05500 (EEL44_05500) | 35558..36229 | + | 672 | WP_024126920 | EAL domain-containing protein | - |
| EEL44_RS05505 (EEL44_05505) | 36250..37233 | + | 984 | WP_241154238 | TraU family protein | traU |
| EEL44_RS05510 (EEL44_05510) | 37349..40147 | + | 2799 | WP_024126922 | conjugal transfer mating pair stabilization protein TraN | traN |
| EEL44_RS05515 (EEL44_05515) | 40186..41046 | - | 861 | WP_000709517 | hypothetical protein | - |
| EEL44_RS05520 (EEL44_05520) | 41169..41810 | - | 642 | WP_024126923 | hypothetical protein | - |
| EEL44_RS05525 (EEL44_05525) | 42103..42423 | + | 321 | WP_024126924 | hypothetical protein | - |
| EEL44_RS05530 (EEL44_05530) | 42727..42912 | + | 186 | WP_001186916 | hypothetical protein | - |
| EEL44_RS05535 (EEL44_05535) | 43131..44099 | + | 969 | WP_024126925 | AAA family ATPase | - |
| EEL44_RS05540 (EEL44_05540) | 44110..45015 | + | 906 | WP_021293644 | hypothetical protein | - |
Host bacterium
| ID | 5053 | GenBank | NZ_CP033514 |
| Plasmid name | pVCR94 | Incompatibility group | IncA/C2 |
| Plasmid size | 171829 bp | Coordinate of oriT [Strand] | 16873..17023 [+] |
| Host baterium | Vibrio cholerae strain E4 |
Cargo genes
| Drug resistance gene | tet(C), tet(B), catA1, aph(3'')-Ib, aph(6)-Id, blaCARB-4, dfrA15, qacE, sul1, sul2 |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |