Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104540 |
| Name | oriT_STLEFF_27|unnamed1 |
| Organism | Shigella flexneri strain STLEFF_27 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP058834 (62630..62981 [+], 352 nt) |
| oriT length | 352 nt |
| IRs (inverted repeats) | 255..262, 271..278 (ACCGCTAG..CTAGCGGT) 192..198, 206..212 (TATAAAA..TTTTATA) 40..47, 50..57 (GCAAAAAC..GTTTTTGC) 4..11, 16..23 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 352 nt
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGAGTTATGAAAAATTAGTTTCTCTTACTCTCTTTATGATATTTAAAAAAGCGGTGTCGGCGCGGCTACAACAACGCGCCGACACCGCTTTGTAGGGGTGGTACTGACTATTTTTATAAAAAACATTATTTTATATTAGGGGTGCTGCTAGCGGCGCGGTGTGTTTTTTTATAGGATACCGCTAGGGGCGCTGCTAGCGGTGCGTCCCTGTTTGCATTATGAATTTTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGTAATCTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file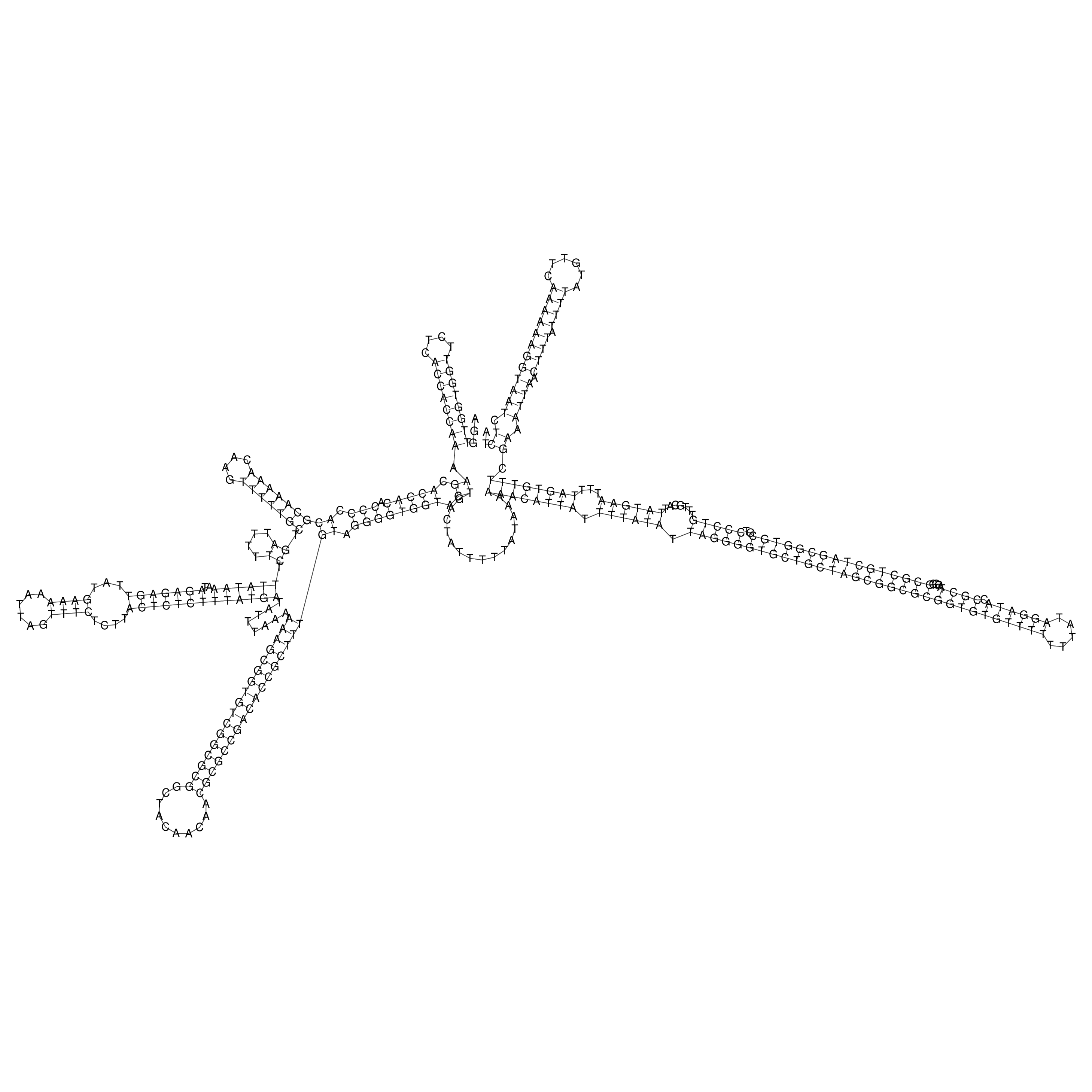
Relaxase
| ID | 3278 | GenBank | WP_000986934 |
| Name | traI_HZT22_RS23870_STLEFF_27|unnamed1 |
UniProt ID | A0A291R9S5 |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 191764.68 Da Isoelectric Point: 5.9593
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGKGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEIRMAEWMQT
LKETGFDIRAYRDAADQRTEIRTQAPGPASQDGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPASLPVSDSPFTALKL
ENGWVETPGHSVSDSATVFASVTQMAMDNATLNGLARSGRDVRLYSSLDETRTAEKLARHPSFTVVSEQI
KARAGETLLETAISLQKAGLHTPAQQAIHLALPVLESKNLAFSMVDLLTEAKSFAAEGTGFTELGGEINA
QIKRGDLLYVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMETLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPASERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTEMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVVIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDIPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWEKNPDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKLMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKPDREVMNAQRLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTELAVRDIAGQERDRSAISERETALPESVLRESQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1466 | GenBank | WP_001151524 |
| Name | WP_001151524_STLEFF_27|unnamed1 |
UniProt ID | A0A5W2DT22 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14517.48 Da Isoelectric Point: 5.0516
MAKVNLYISNDAYEKINAIIEKRRQEGAREKDVSFSATASMLLELGLRVHEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5W2DT22 |
| ID | 1467 | GenBank | WP_001369361 |
| Name | WP_001369361_STLEFF_27|unnamed1 |
UniProt ID | B7LI64 |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15409.70 Da Isoelectric Point: 9.9240
MLLKRFGTRSATGKMVKLKLPVDVESLLIEASNRSGRSRSFEAVIRLKDHLHRYPKFNRAGNIYGKSLVK
YLTMRLDDETNQLLIAAKNRSGWCKTDEAADRVIDHLIKFPDFYNSEIFREADKEEDITFNTL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B7LI64 |
T4CP
| ID | 3212 | GenBank | WP_113421867 |
| Name | traC_HZT22_RS23775_STLEFF_27|unnamed1 |
UniProt ID | _ |
| Length | 875 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 875 a.a. Molecular weight: 99470.50 Da Isoelectric Point: 6.6265
MNNPLEAVTQAVNSLVTALKLPDESAKANEVLGEMSFPQFSRLLPYRDYNQESGLFMNDTTMGFMLEAIP
INGANESIVEALDHMLRTKLPRGIPLCIHLMSSQLVGDRIEYGLREFSWSGEQAERFNAITRAYYMKAAA
TQFPLPEGMNLPLTLRHYRVFISYCSPSKKKSRADILEMENLVKIIRASFHGAKITTQTVDAQAFIEIVG
EMINHNPDSLYPKRRQLDPYSDLNYQCVEDSFDLKVRADYLTLGLRENGRNSTARILNFHLARNPEIAFL
WNMADNYSNLLNPEMSISCPFILTLTLVVEDQVKTHSEANLKYMDLEKKSKTSYAKWFPSVEKEAKEWGE
LRQRLGSGQSSVVSYFLNITAFCKDNNETALEVEQDILNSFRKNGFELISPRFNHMRNFLTCLPFMAGKG
LFKQLKEAGVVQRAESFNVANLMPLVADNPLTPAGLLAPTYRNQLAFIDIFFKGMNNTNYNMAVCGTSGA
GKTGLIQPLIRSVLDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLRFNPFANITDIDQSAERVRDQLSV
MASPNGNLDEVHEGLLLQAVRASWLAKKKQARIDDVVDFLKNARDNDQYVESPTIRSRLDEMIVLLDQYT
ANGTYGRYFNSDEPSLRDDAKMVVLELGGLEDRPSLLVAVMFSLIIYIENRMYRTPRTLKKLNVIDEGWR
LLDFKNRKVGEFIQKGYRTCRRHTGAYITITQNIVDFDSDKASSAARAAWGNSSYKIILKQSAKEFAKYN
QLFPDQFQPLQRDMIGKFGAAKDLWFSSFLLQVENHSSWHRLFVDPLSRAMYSSDGPDFEFVQQKRREGM
SIHEAVWQLAWKKSGPEMASLEAWLEEHEKYRSVA
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3213 | GenBank | WP_000009360 |
| Name | traD_HZT22_RS23865_STLEFF_27|unnamed1 |
UniProt ID | _ |
| Length | 732 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 732 a.a. Molecular weight: 83294.06 Da Isoelectric Point: 5.1048
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLASVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVSTGKSEVIRRLANYAR
KRGDMVVIYDRSCEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNVANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVVSMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVLNTRAFFRSPSHQIA
EFAAGEIGEKEHLKASLQYSYGADPVRDGISTGKEMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPVSPAINDKKSDSGVNIPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQEN
HPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 61989..84893
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HZT22_RS23655 (HZT22_23515) | 57955..58389 | + | 435 | WP_000845907 | conjugation system SOS inhibitor PsiB | - |
| HZT22_RS23660 (HZT22_23520) | 58386..59105 | + | 720 | WP_001276240 | plasmid SOS inhibition protein A | - |
| HZT22_RS23665 (HZT22_23525) | 59117..59305 | - | 189 | WP_001299721 | hypothetical protein | - |
| HZT22_RS23670 | 59327..59476 | + | 150 | Protein_77 | plasmid maintenance protein Mok | - |
| HZT22_RS23675 (HZT22_23530) | 59418..59543 | + | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| HZT22_RS23680 (HZT22_23535) | 59763..59993 | + | 231 | WP_071845785 | hypothetical protein | - |
| HZT22_RS23685 | 59991..60164 | - | 174 | Protein_80 | hypothetical protein | - |
| HZT22_RS23690 (HZT22_23540) | 60234..60440 | + | 207 | WP_063609967 | hypothetical protein | - |
| HZT22_RS23695 (HZT22_23545) | 60465..60752 | + | 288 | WP_063072995 | hypothetical protein | - |
| HZT22_RS23700 (HZT22_23550) | 60871..61692 | + | 822 | WP_001241365 | DUF932 domain-containing protein | - |
| HZT22_RS23705 (HZT22_23555) | 61989..62579 | - | 591 | WP_222908884 | transglycosylase SLT domain-containing protein | virB1 |
| HZT22_RS23710 (HZT22_23560) | 62982..63365 | + | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HZT22_RS23715 (HZT22_23565) | 63552..64241 | + | 690 | WP_000283379 | conjugal transfer transcriptional regulator TraJ | - |
| HZT22_RS23720 (HZT22_23570) | 64340..64735 | + | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| HZT22_RS23725 (HZT22_23575) | 64768..65133 | + | 366 | WP_000994779 | type IV conjugative transfer system pilin TraA | - |
| HZT22_RS23730 (HZT22_23580) | 65148..65459 | + | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HZT22_RS23735 (HZT22_23585) | 65481..66047 | + | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HZT22_RS23740 (HZT22_23590) | 66034..66762 | + | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| HZT22_RS23745 (HZT22_23595) | 66762..68189 | + | 1428 | WP_000146638 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HZT22_RS23750 (HZT22_23600) | 68179..68763 | + | 585 | WP_000002795 | conjugal transfer pilus-stabilizing protein TraP | - |
| HZT22_RS23755 (HZT22_23605) | 68750..69070 | + | 321 | WP_001057302 | conjugal transfer protein TrbD | virb4 |
| HZT22_RS23760 (HZT22_23610) | 69063..69314 | + | 252 | WP_001038341 | conjugal transfer protein TrbG | - |
| HZT22_RS23765 (HZT22_23615) | 69311..69826 | + | 516 | WP_000809893 | type IV conjugative transfer system lipoprotein TraV | traV |
| HZT22_RS23770 (HZT22_23620) | 69961..70182 | + | 222 | WP_072651814 | conjugal transfer protein TraR | - |
| HZT22_RS23775 (HZT22_23625) | 70342..72969 | + | 2628 | WP_113421867 | type IV secretion system protein TraC | virb4 |
| HZT22_RS23780 (HZT22_23630) | 72966..73352 | + | 387 | WP_000214082 | type-F conjugative transfer system protein TrbI | - |
| HZT22_RS23785 (HZT22_23635) | 73349..73981 | + | 633 | WP_001203720 | type-F conjugative transfer system protein TraW | traW |
| HZT22_RS23790 (HZT22_23640) | 73978..74970 | + | 993 | WP_000830838 | conjugal transfer pilus assembly protein TraU | traU |
| HZT22_RS23795 (HZT22_23645) | 75000..75305 | + | 306 | WP_000224411 | hypothetical protein | - |
| HZT22_RS23800 (HZT22_23650) | 75314..75952 | + | 639 | WP_001080257 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HZT22_RS23805 (HZT22_23655) | 75949..77757 | + | 1809 | WP_000821863 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HZT22_RS23810 (HZT22_23660) | 77784..78041 | + | 258 | WP_000864320 | conjugal transfer protein TrbE | - |
| HZT22_RS23815 (HZT22_23665) | 78034..78777 | + | 744 | WP_001030371 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HZT22_RS23820 (HZT22_23670) | 78793..79131 | + | 339 | WP_001287905 | conjugal transfer protein TrbA | - |
| HZT22_RS23825 (HZT22_23675) | 79258..79542 | + | 285 | WP_001448202 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HZT22_RS23830 (HZT22_23680) | 79529..80074 | + | 546 | WP_000059824 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HZT22_RS23835 (HZT22_23685) | 80004..80344 | + | 341 | Protein_110 | P-type conjugative transfer protein TrbJ | - |
| HZT22_RS23840 (HZT22_23690) | 80325..80717 | + | 393 | WP_000660703 | F-type conjugal transfer protein TrbF | - |
| HZT22_RS23845 (HZT22_23695) | 80704..82077 | + | 1374 | WP_112901602 | conjugal transfer pilus assembly protein TraH | traH |
| HZT22_RS23850 (HZT22_23700) | 82074..84893 | + | 2820 | WP_113421866 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HZT22_RS23855 (HZT22_23705) | 84912..85409 | + | 498 | WP_000605861 | hypothetical protein | - |
| HZT22_RS23860 (HZT22_23710) | 85441..86172 | + | 732 | WP_000850422 | conjugal transfer complement resistance protein TraT | - |
| HZT22_RS23865 (HZT22_23715) | 86425..88623 | + | 2199 | WP_000009360 | type IV conjugative transfer system coupling protein TraD | - |
Host bacterium
| ID | 4979 | GenBank | NZ_CP058834 |
| Plasmid name | STLEFF_27|unnamed1 | Incompatibility group | IncFII |
| Plasmid size | 98962 bp | Coordinate of oriT [Strand] | 62630..62981 [+] |
| Host baterium | Shigella flexneri strain STLEFF_27 |
Cargo genes
| Drug resistance gene | cmlA1, qacE, blaCTX-M-14, erm(B), mph(A) |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |