Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104524 |
| Name | oriT_SWHIN_101|unnamed2 |
| Organism | Shigella flexneri strain SWHIN_101 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055115 (31588..31660 [-], 73 nt) |
| oriT length | 73 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 73 nt
>oriT_SWHIN_101|unnamed2
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
GTCGGGGCAAAGCCCTGACCAGGTGGGGAATGTCTGAGTGCGCGTGCGCGGTCCGACATTCCCACATCCTGTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file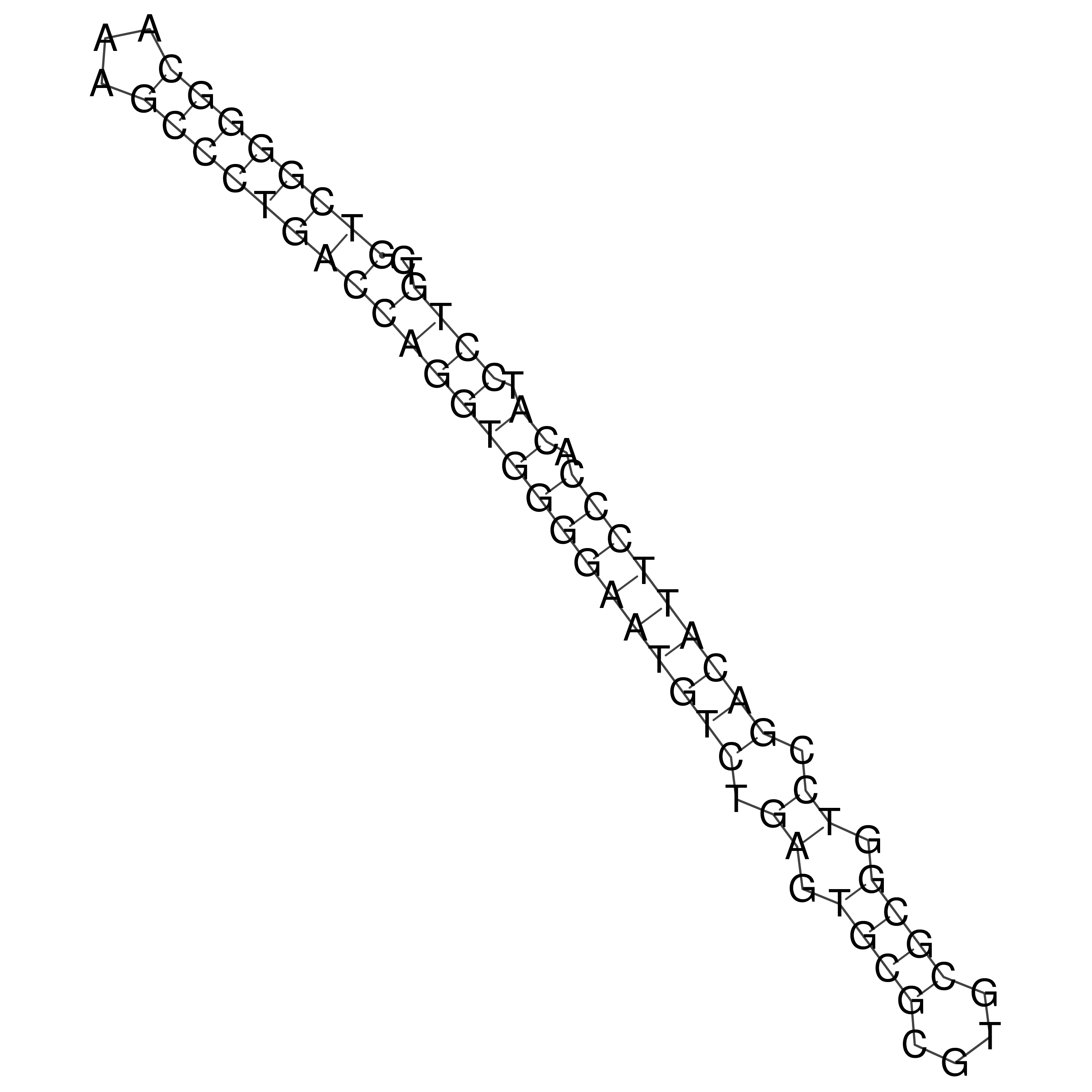
Relaxase
| ID | 3272 | GenBank | WP_058100850 |
| Name | Relaxase_HUZ81_RS24980_SWHIN_101|unnamed2 |
UniProt ID | A0A1M0DNA5 |
| Length | 906 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 906 a.a. Molecular weight: 105098.51 Da Isoelectric Point: 8.3663
>WP_058100850.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterobacteriaceae]
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWHSVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQLRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
MNAIIPHKRRDGRSSFEDLIAYTSVRDDVQENELTEDSEIKSDVPHRNRFRRLVDYATRLRNEKFVSLID
VMKDGSQWVNFYGVTCFHNCNSLETAAEEMQYKADKAVFSRNDTDPVFHYILSWPAHESPRPEQLFDCVR
HTLKSLELSKHQYVAAVHTDTDNLHVHVAVNRVHPETGYINWLSWSQEKLSRACRELELKHGFAPDNGCF
VHAPGNRIVRKTALVRERRNAWRRGKKQTFREYIAQMSIAGLREEPAQDWLSLHKRLASDGLYITMQEGE
LVVKDGWDRAREGVALSSFGPSWTAEKLGRKLGEYQPVPTDIFSQVGTPGRYDPEAINVDIRPEKVAETE
SLKQYACRHFAERLPAMARNGELKSCLAVHRTLAEAGLWMGIQHGHLVLHDGFDKQQTPVRADSVWPLMT
LDYMQDLDGGWHSVPKDIFTQVIPGERFRGHSIGTQAVSDYEWYRMRMGTGPQGAIKRELFSDKESLWGY
ALIQCRSQIEEMIAGGDFSWQACHEMFARKGLMLQKQHHGLVIVDAFNHELTPVKASSIHPDLTLSRAEP
QAGPFEIAAADIFERVKPECRYNPELAASDEVEPGFRRDPELRRERREARAAAREDLRARYLAWKEHWRK
PDLRYGERLREIHAACRRRKAHIRVQFRDPQLRKLHYHIAEVQRMQALIRLKENVREERLALMAEGKWYP
LSYRQWVEQQAAQGDRAAVSQLRGWDYRDRRSRNKDKRRTTNADRCVVLCEPGGTPLFNNVAKLEARLQK
NGSVHFRDTRTGKNVCTDYGDRVVFYHHTDRNELAEKLDLIAPVLFSRNGKLGFEPEGSYQQFNDVFAEM
VAWHNAAGITGNGHFTITRPDVDLHRQRSEQYYREYIRQQTRLSESHDDNYTLRQEKTWEPPSPGM
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A1M0DNA5 |
Auxiliary protein
| ID | 1462 | GenBank | WP_001291058 |
| Name | WP_001291058_SWHIN_101|unnamed2 |
UniProt ID | A0A7I0L241 |
| Length | 110 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 110 a.a. Molecular weight: 12730.52 Da Isoelectric Point: 9.9656
>WP_001291058.1 MULTISPECIES: plasmid mobilization protein MobA [Enterobacteriaceae]
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
MSEKKTRTGSENRKRIVKFTARFTEDEAEIVREKAESSGQTVSTFIRSSSLDKVVNCRTDDRMIDEIMRL
GRLQKHLFVEGKRTGDKEYAEVLVAITEYVNALRRDLMGR
Protein domains
No domain identified.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A7I0L241 |
T4CP
| ID | 3202 | GenBank | WP_058100849 |
| Name | trbC_HUZ81_RS24985_SWHIN_101|unnamed2 |
UniProt ID | _ |
| Length | 766 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 766 a.a. Molecular weight: 87195.08 Da Isoelectric Point: 6.6676
>WP_058100849.1 MULTISPECIES: F-type conjugative transfer protein TrbC [Enterobacteriaceae]
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVRERKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
MSQHHVNPDLIHRTAWGNPLWNALHNLNIIGLCLAGSIITALIWPLALPVCLLFTLVTSVIFTLQRWRCP
LRMPMTLSLDDPSQDRKVRRSLFSFWPTLFQYEADETSPARGIFYVGYRRINDIGRELWLSMDDLTRHII
FFSTTGGGKTETTFAWLLNPLCWGRGFTFVDGKAQNDTTRTIWYLSRRFGREDDIEVINFMNGGKSRSEI
IQSGEKSRPQSNTWNPFAFSTEAFTAETMQSMLPQNVQGGEWQSRAIAMNKALVFGTKFWCVRERKTMSL
QMLREHMTLEGMAKLYCRGLDDQWPEEAIAPLRNYLQDVPGFDMSLVRTPSAWTEEPRKQHAYLSGQFSE
TFTTFAETFGDVFAADAGDIDIRDSIHSDRILIVMIPALDTSAHTTSALGRMFITQQSMILARDLGYRLE
GTDAQTLEVKKYKGRFPYICILDEVGAYYTDRIAVEATQVRSLEFSLIMTAQDQERIEGQTSATNTATLM
QNAGTKFAGRIVSDDKTARTVKNAAGEEARARMGSLQRHDGVMGESWVDGNQITIQMESKIDVQDLIRLN
AGEFFTVFQGDVVPSASVYIPDSEKSCDSDPVVINRYIAIEAPRLERLRRLVPRTVQRRLPTPEHVSSII
GVLTEKTSRKRRKKHTEPYRIIDTFQDQLKRTQTSLDLLPQYDTDIESRANELWKKAVHTINNTTREERR
VCYITLNRPEEEHSGPEDIPSVKAILNTLLPLEILLPVAENNKSQTINNDHPQKSCGAQPTTDEKY
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 37246..81558
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HUZ81_RS24985 (HUZ81_24750) | 34965..37265 | - | 2301 | WP_058100849 | F-type conjugative transfer protein TrbC | - |
| HUZ81_RS24990 (HUZ81_24755) | 37246..38370 | - | 1125 | WP_058100848 | DsbC family protein | trbB |
| HUZ81_RS24995 (HUZ81_24760) | 38367..39629 | - | 1263 | WP_045145416 | IncI1-type conjugal transfer protein TrbA | trbA |
| HUZ81_RS25000 (HUZ81_24765) | 40294..40761 | - | 468 | WP_045145415 | thermonuclease family protein | - |
| HUZ81_RS25005 (HUZ81_24770) | 40926..41549 | + | 624 | WP_045145414 | helix-turn-helix domain-containing protein | - |
| HUZ81_RS25010 (HUZ81_24775) | 41747..41962 | - | 216 | WP_000266543 | hypothetical protein | - |
| HUZ81_RS25015 (HUZ81_24780) | 41966..42334 | - | 369 | WP_001302696 | hypothetical protein | - |
| HUZ81_RS25020 (HUZ81_24785) | 42346..42537 | - | 192 | WP_000751583 | hypothetical protein | - |
| HUZ81_RS25025 (HUZ81_24790) | 42634..42876 | - | 243 | WP_000650933 | hypothetical protein | - |
| HUZ81_RS25030 (HUZ81_24795) | 43205..43357 | + | 153 | WP_021532728 | Hok/Gef family protein | - |
| HUZ81_RS25035 (HUZ81_24800) | 43926..44078 | + | 153 | WP_001367784 | Hok/Gef family protein | - |
| HUZ81_RS25040 (HUZ81_24805) | 44075..44236 | - | 162 | WP_021580032 | hypothetical protein | - |
| HUZ81_RS25045 (HUZ81_24810) | 44298..45869 | - | 1572 | WP_239622816 | IS66-like element ISCro1 family transposase | - |
| HUZ81_RS25050 (HUZ81_24815) | 45889..46236 | - | 348 | WP_012904570 | IS66 family insertion sequence element accessory protein TnpB | - |
| HUZ81_RS25055 (HUZ81_24820) | 46236..46913 | - | 678 | WP_053287942 | IS66-like element accessory protein TnpA | - |
| HUZ81_RS25060 | 47099..47230 | - | 132 | Protein_62 | ethanolamine utilization protein EutE | - |
| HUZ81_RS25065 (HUZ81_24825) | 47325..47534 | + | 210 | WP_000062946 | HEAT repeat domain-containing protein | - |
| HUZ81_RS25070 (HUZ81_24830) | 47599..48246 | - | 648 | WP_072643575 | plasmid IncI1-type surface exclusion protein ExcA | - |
| HUZ81_RS25075 (HUZ81_24835) | 48334..50493 | - | 2160 | WP_058100846 | DotA/TraY family protein | traY |
| HUZ81_RS25080 (HUZ81_24840) | 50566..51135 | - | 570 | WP_058100845 | conjugal transfer protein TraX | - |
| HUZ81_RS25085 (HUZ81_24845) | 51132..52337 | - | 1206 | WP_052908626 | conjugal transfer protein TraW | traW |
| HUZ81_RS25090 (HUZ81_24850) | 52298..52915 | - | 618 | WP_052908627 | hypothetical protein | traV |
| HUZ81_RS25095 (HUZ81_24855) | 52915..55959 | - | 3045 | WP_058100844 | conjugal transfer protein | traU |
| HUZ81_RS25100 (HUZ81_24860) | 56135..56515 | - | 381 | WP_058100843 | hypothetical protein | - |
| HUZ81_RS25105 (HUZ81_24865) | 56515..56874 | - | 360 | WP_058100842 | hypothetical protein | - |
| HUZ81_RS25110 (HUZ81_24870) | 56931..57749 | - | 819 | WP_239622807 | conjugal transfer protein TraT | traT |
| HUZ81_RS25520 (HUZ81_24875) | 57664..57915 | - | 252 | WP_001277575 | hypothetical protein | - |
| HUZ81_RS25120 (HUZ81_24880) | 57972..58370 | - | 399 | WP_021532718 | DUF6750 family protein | traR |
| HUZ81_RS25125 (HUZ81_24885) | 58417..58947 | - | 531 | WP_227316726 | conjugal transfer protein TraQ | traQ |
| HUZ81_RS25130 (HUZ81_24890) | 58944..59657 | - | 714 | WP_072657236 | conjugal transfer protein TraP | traP |
| HUZ81_RS25135 (HUZ81_24895) | 59654..60985 | - | 1332 | WP_058100840 | conjugal transfer protein TraO | traO |
| HUZ81_RS25140 (HUZ81_24900) | 60989..61963 | - | 975 | WP_021532715 | DotH/IcmK family type IV secretion protein | traN |
| HUZ81_RS25145 (HUZ81_24905) | 61974..62669 | - | 696 | WP_029391932 | DotI/IcmL family type IV secretion protein | traM |
| HUZ81_RS25150 (HUZ81_24910) | 62681..63031 | - | 351 | WP_021532713 | hypothetical protein | traL |
| HUZ81_RS25155 (HUZ81_24915) | 63048..67109 | - | 4062 | WP_058100839 | LPD7 domain-containing protein | - |
| HUZ81_RS25160 (HUZ81_24920) | 67173..67463 | - | 291 | WP_000817799 | hypothetical protein | traK |
| HUZ81_RS25165 (HUZ81_24925) | 67460..68608 | - | 1149 | WP_058100838 | plasmid transfer ATPase TraJ | virB11 |
| HUZ81_RS25170 (HUZ81_24930) | 68592..69428 | - | 837 | WP_000745049 | type IV secretory system conjugative DNA transfer family protein | traI |
| HUZ81_RS25175 (HUZ81_24935) | 69425..69883 | - | 459 | WP_001170111 | DotD/TraH family lipoprotein | - |
| HUZ81_RS25180 (HUZ81_24940) | 69987..71189 | - | 1203 | WP_053287757 | conjugal transfer protein TraF | - |
| HUZ81_RS25185 (HUZ81_24945) | 71291..72112 | - | 822 | WP_001681675 | hypothetical protein | traE |
| HUZ81_RS25190 (HUZ81_24950) | 72386..72706 | + | 321 | WP_000695052 | hypothetical protein | - |
| HUZ81_RS25195 (HUZ81_24955) | 72798..73088 | - | 291 | WP_021532709 | hypothetical protein | - |
| HUZ81_RS25200 (HUZ81_24960) | 73130..74299 | - | 1170 | WP_021532708 | site-specific integrase | - |
| HUZ81_RS25205 (HUZ81_24965) | 74969..75208 | + | 240 | WP_000271302 | hypothetical protein | - |
| HUZ81_RS25210 (HUZ81_24970) | 75213..75602 | - | 390 | Protein_92 | phage tail protein | - |
| HUZ81_RS25215 (HUZ81_24975) | 75886..77163 | - | 1278 | WP_077891451 | shufflon system plasmid conjugative transfer pilus tip adhesin PilV | - |
| HUZ81_RS25220 (HUZ81_24980) | 77160..77867 | - | 708 | WP_152920992 | A24 family peptidase | - |
| HUZ81_RS25225 (HUZ81_24985) | 77827..78303 | - | 477 | WP_029391914 | lytic transglycosylase domain-containing protein | virB1 |
| HUZ81_RS25230 (HUZ81_24990) | 78362..78898 | - | 537 | WP_058100836 | type 4 pilus major pilin | - |
| HUZ81_RS25235 (HUZ81_24995) | 78948..80048 | - | 1101 | WP_001696153 | type II secretion system F family protein | - |
| HUZ81_RS25240 (HUZ81_25000) | 80050..81558 | - | 1509 | WP_058100835 | ATPase, T2SS/T4P/T4SS family | virB11 |
| HUZ81_RS25245 (HUZ81_25005) | 81655..82329 | - | 675 | WP_226256164 | hypothetical protein | - |
| HUZ81_RS25250 (HUZ81_25010) | 82347..82901 | - | 555 | WP_021532699 | hypothetical protein | - |
| HUZ81_RS25255 (HUZ81_25015) | 82950..83402 | - | 453 | WP_021532698 | type IV pilus biogenesis protein PilP | - |
| HUZ81_RS25260 (HUZ81_25020) | 83392..84687 | - | 1296 | WP_058100834 | type 4b pilus protein PilO2 | - |
| HUZ81_RS25265 (HUZ81_25025) | 84708..86327 | - | 1620 | WP_021532696 | PilN family type IVB pilus formation outer membrane protein | - |
Host bacterium
| ID | 4963 | GenBank | NZ_CP055115 |
| Plasmid name | SWHIN_101|unnamed2 | Incompatibility group | IncB/O/K/Z |
| Plasmid size | 91560 bp | Coordinate of oriT [Strand] | 31588..31660 [-] |
| Host baterium | Shigella flexneri strain SWHIN_101 |
Cargo genes
| Drug resistance gene | aph(3')-IIa |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |