Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104521 |
| Name | oriT_SWHEFF_71|unnamed1 |
| Organism | Shigella flexneri strain SWHEFF_71 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055016 (155968..156319 [-], 352 nt) |
| oriT length | 352 nt |
| IRs (inverted repeats) | 255..262, 271..278 (ACCGCTAG..CTAGCGGT) 192..198, 206..212 (TATAAAA..TTTTATA) 40..47, 50..57 (GCAAAAAC..GTTTTTGC) 4..11, 16..23 (TTGGTGGT..ACCACCAA) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 352 nt
AGGTTGGTGGTTCTCACCACCAAAAGCACCACACCCCACGCAAAAACAAGTTTTTGCTGATTTTTCTTTATAAATAGAGAGTTATGAAAAATTAGTTTCTCTTACTCTCTTTATGATATTTAAAAAAGCGGTGTCGGCGCGGCTACAACAACGCGCCGACACCGCTTTGTAGGGGTGGTACTGACTATTTTTATAAAAAACATTATTTTATATTAGGGGTGCTGCTAGCGGCGCGGTGTGTTTTTTTATAGGATACCGCTAGGGGCGCTGCTAGCGGTGCGTCCCTGTTTGCATTATGAATTTTAGTGTTTCGAAATTAACTTTATTTTATGTTCAAAAAAGGTAATCTCTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file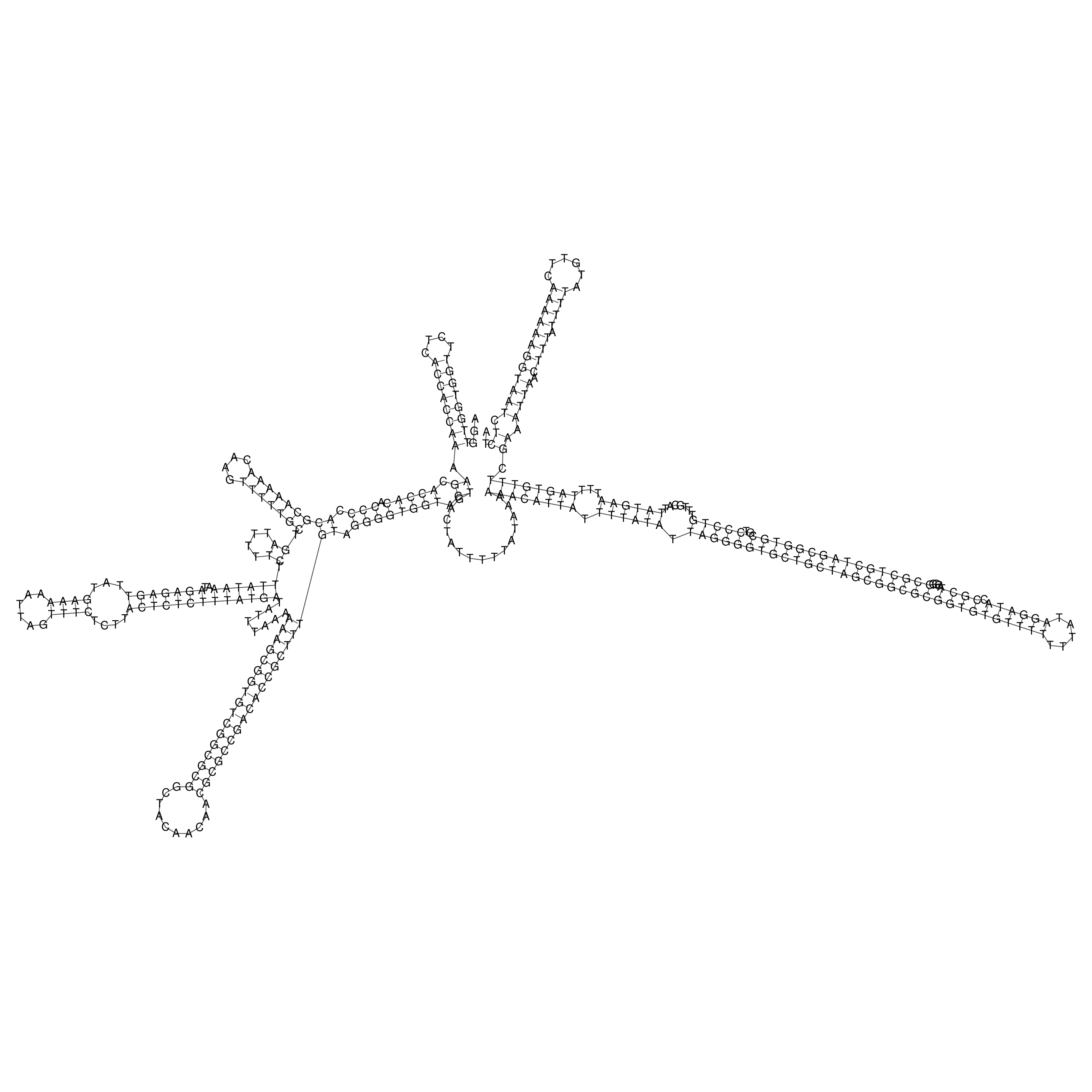
Relaxase
| ID | 3270 | GenBank | WP_096942428 |
| Name | traI_HUZ78_RS25075_SWHEFF_71|unnamed1 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192163.81 Da Isoelectric Point: 6.0655
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGRGAEQLGLQGSVDKDVFTRLLEGRLPDGADLSRMQ
DGSNKHRPGYDLTFSAPKSVSMMAMLGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNMV
MALFNHDTSRDQEPQLHTHAVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQIAFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQAIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMAEWMQT
LKETGFDIRAYRDAADQRAETRTQAPGAVSQEGPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPE
NGVIERARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVTVHPEKSVPRTAGYS
DAVSVLAQDRPSLAIVSGQGGAVGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLLEGMAFTPGSTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPVTIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHPEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVSGGDRLQVASVSEDAMTVVVPGRAEPATLPVSDSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNGTLNGLARSGRDVRLYSSLDEPRTAEKLARHPSFTVVSEQI
KTRAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFAAEGTSFADLGREIDA
QIKRGDLLHVDVAKGYGTDLLVSCASYEAEKSILRHILEGKEAVTPLMERVPVELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRQEGAWAPEHSVTEFSHSQEAKLAEA
QQKAMQKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALALVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDKIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQKTRVIRPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVL
EPKSDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGRSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAENKPDLP
DGKTEQAVREIAGQERDRSAISEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1458 | GenBank | WP_001369361 |
| Name | WP_001369361_SWHEFF_71|unnamed1 |
UniProt ID | B7LI64 |
| Length | 133 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 133 a.a. Molecular weight: 15409.70 Da Isoelectric Point: 9.9240
MLLKRFGTRSATGKMVKLKLPVDVESLLIEASNRSGRSRSFEAVIRLKDHLHRYPKFNRAGNIYGKSLVK
YLTMRLDDETNQLLIAAKNRSGWCKTDEAADRVIDHLIKFPDFYNSEIFREADKEEDITFNTL
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | B7LI64 |
| ID | 1459 | GenBank | WP_001151524 |
| Name | WP_001151524_SWHEFF_71|unnamed1 |
UniProt ID | A0A5W2DT22 |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14517.48 Da Isoelectric Point: 5.0516
MAKVNLYISNDAYEKINAIIEKRRQEGAREKDVSFSATASMLLELGLRVHEAQMERKESAFNQTEFNKLL
LECVVKTQSSVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A5W2DT22 |
T4CP
| ID | 3200 | GenBank | WP_022630316 |
| Name | traD_HUZ78_RS25080_SWHEFF_71|unnamed1 |
UniProt ID | _ |
| Length | 738 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 738 a.a. Molecular weight: 83920.60 Da Isoelectric Point: 5.0524
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTENPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGESFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKAAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASEQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPRDINPEMENRLSAVLAAREAEGRQMASLFEPDVPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPQQPQQPQQPVSPAINDKKSDSGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYE
AWQQENHPDIQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 128798..156890
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HUZ78_RS25080 (HUZ78_24900) | 128798..131014 | - | 2217 | WP_022630316 | type IV conjugative transfer system coupling protein TraD | virb4 |
| HUZ78_RS25085 (HUZ78_24905) | 131268..131999 | - | 732 | WP_000850424 | conjugal transfer complement resistance protein TraT | - |
| HUZ78_RS25090 (HUZ78_24910) | 132013..132522 | - | 510 | WP_000628099 | conjugal transfer entry exclusion protein TraS | - |
| HUZ78_RS25095 (HUZ78_24915) | 132519..135344 | - | 2826 | WP_195715507 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HUZ78_RS25100 (HUZ78_24920) | 135341..136714 | - | 1374 | WP_001137358 | conjugal transfer pilus assembly protein TraH | traH |
| HUZ78_RS25105 (HUZ78_24925) | 136714..137076 | - | 363 | WP_010379880 | P-type conjugative transfer protein TrbJ | - |
| HUZ78_RS25110 (HUZ78_24930) | 137006..137551 | - | 546 | WP_000052614 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HUZ78_RS25115 (HUZ78_24935) | 137538..137822 | - | 285 | WP_000624110 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HUZ78_RS25120 (HUZ78_24940) | 137941..138288 | - | 348 | WP_001287893 | conjugal transfer protein TrbA | - |
| HUZ78_RS25125 (HUZ78_24945) | 138304..139047 | - | 744 | WP_001030395 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HUZ78_RS25130 (HUZ78_24950) | 139040..139297 | - | 258 | WP_000864311 | conjugal transfer protein TrbE | - |
| HUZ78_RS25135 (HUZ78_24955) | 139324..141132 | - | 1809 | WP_089645794 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HUZ78_RS25140 (HUZ78_24960) | 141129..141767 | - | 639 | WP_122994733 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HUZ78_RS25145 (HUZ78_24965) | 141795..142247 | - | 453 | WP_000069428 | hypothetical protein | - |
| HUZ78_RS25150 (HUZ78_24970) | 142277..142738 | - | 462 | WP_000277841 | hypothetical protein | - |
| HUZ78_RS25155 (HUZ78_24975) | 142764..143756 | - | 993 | WP_000830839 | conjugal transfer pilus assembly protein TraU | traU |
| HUZ78_RS25160 (HUZ78_24980) | 143753..144385 | - | 633 | WP_001203722 | type-F conjugative transfer system protein TraW | traW |
| HUZ78_RS25165 (HUZ78_24985) | 144382..144768 | - | 387 | WP_000214094 | type-F conjugative transfer system protein TrbI | - |
| HUZ78_RS25170 (HUZ78_24990) | 144765..147392 | - | 2628 | WP_205850319 | type IV secretion system protein TraC | virb4 |
| HUZ78_RS25175 (HUZ78_24995) | 147552..147773 | - | 222 | WP_001278685 | conjugal transfer protein TraR | - |
| HUZ78_RS25180 (HUZ78_25000) | 147908..148423 | - | 516 | WP_000809841 | type IV conjugative transfer system lipoprotein TraV | traV |
| HUZ78_RS25185 (HUZ78_25005) | 148420..148671 | - | 252 | WP_001038342 | conjugal transfer protein TrbG | - |
| HUZ78_RS25190 (HUZ78_25010) | 148698..148829 | - | 132 | Protein_160 | conjugal transfer protein TrbD | - |
| HUZ78_RS25200 | 150113..150241 | - | 129 | Protein_162 | conjugal transfer protein TrbD | - |
| HUZ78_RS25205 (HUZ78_25020) | 150180..150770 | - | 591 | WP_000002778 | conjugal transfer pilus-stabilizing protein TraP | - |
| HUZ78_RS25210 (HUZ78_25025) | 150760..152187 | - | 1428 | WP_000146649 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HUZ78_RS25215 (HUZ78_25030) | 152187..152915 | - | 729 | WP_001230787 | type-F conjugative transfer system secretin TraK | traK |
| HUZ78_RS25220 (HUZ78_25035) | 152902..153468 | - | 567 | WP_000399792 | type IV conjugative transfer system protein TraE | traE |
| HUZ78_RS25225 (HUZ78_25040) | 153490..153801 | - | 312 | WP_000012106 | type IV conjugative transfer system protein TraL | traL |
| HUZ78_RS25230 (HUZ78_25045) | 153816..154181 | - | 366 | WP_000994779 | type IV conjugative transfer system pilin TraA | - |
| HUZ78_RS25235 (HUZ78_25050) | 154214..154609 | - | 396 | WP_001309237 | conjugal transfer relaxosome DNA-bindin protein TraY | - |
| HUZ78_RS25240 (HUZ78_25055) | 154708..155397 | - | 690 | WP_000283379 | conjugal transfer transcriptional regulator TraJ | - |
| HUZ78_RS25245 (HUZ78_25060) | 155584..155967 | - | 384 | WP_001151524 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HUZ78_RS25250 (HUZ78_25065) | 156300..156890 | + | 591 | WP_222908884 | transglycosylase SLT domain-containing protein | virB1 |
| HUZ78_RS25255 (HUZ78_25070) | 157187..158008 | - | 822 | WP_001241365 | DUF932 domain-containing protein | - |
| HUZ78_RS25260 (HUZ78_25075) | 158128..158415 | - | 288 | WP_000107535 | hypothetical protein | - |
| HUZ78_RS25265 (HUZ78_25080) | 158440..158646 | - | 207 | WP_033804333 | hypothetical protein | - |
| HUZ78_RS25270 | 158716..158889 | + | 174 | Protein_176 | hypothetical protein | - |
| HUZ78_RS25275 | 158887..159117 | - | 231 | WP_071845785 | hypothetical protein | - |
| HUZ78_RS25280 (HUZ78_25090) | 159337..159462 | - | 126 | WP_001372321 | type I toxin-antitoxin system Hok family toxin | - |
| HUZ78_RS25285 | 159404..159553 | - | 150 | Protein_179 | plasmid maintenance protein Mok | - |
| HUZ78_RS25290 (HUZ78_25095) | 159575..159763 | + | 189 | WP_001299721 | hypothetical protein | - |
| HUZ78_RS25295 (HUZ78_25100) | 159732..160494 | - | 763 | Protein_181 | plasmid SOS inhibition protein A | - |
| HUZ78_RS25300 (HUZ78_25105) | 160491..160925 | - | 435 | WP_000845895 | conjugation system SOS inhibitor PsiB | - |
Host bacterium
| ID | 4960 | GenBank | NZ_CP055016 |
| Plasmid name | SWHEFF_71|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 183928 bp | Coordinate of oriT [Strand] | 155968..156319 [-] |
| Host baterium | Shigella flexneri strain SWHEFF_71 |
Cargo genes
| Drug resistance gene | tet(A), aadA2, dfrA12, aac(3)-IId, blaTEM-1B, aph(6)-Id, aph(3'')-Ib, sul2, sitABCD |
| Virulence gene | exeG, exeE, stcE |
| Metal resistance gene | merR, merT, merP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |