Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104408 |
| Name | oriT_RHBSTW-00405|unnamed |
| Organism | Klebsiella pneumoniae strain RHBSTW-00405 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP055520 (71783..71831 [-], 49 nt) |
| oriT length | 49 nt |
| IRs (inverted repeats) | 6..13, 16..23 (GCAAAATT..AATTTTGC) |
| Location of nic site | 32..33 |
| Conserved sequence flanking the nic site |
TGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 49 nt
>oriT_RHBSTW-00405|unnamed
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
AATCTGCAAAATTTTAATTTTGCGTAGTGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file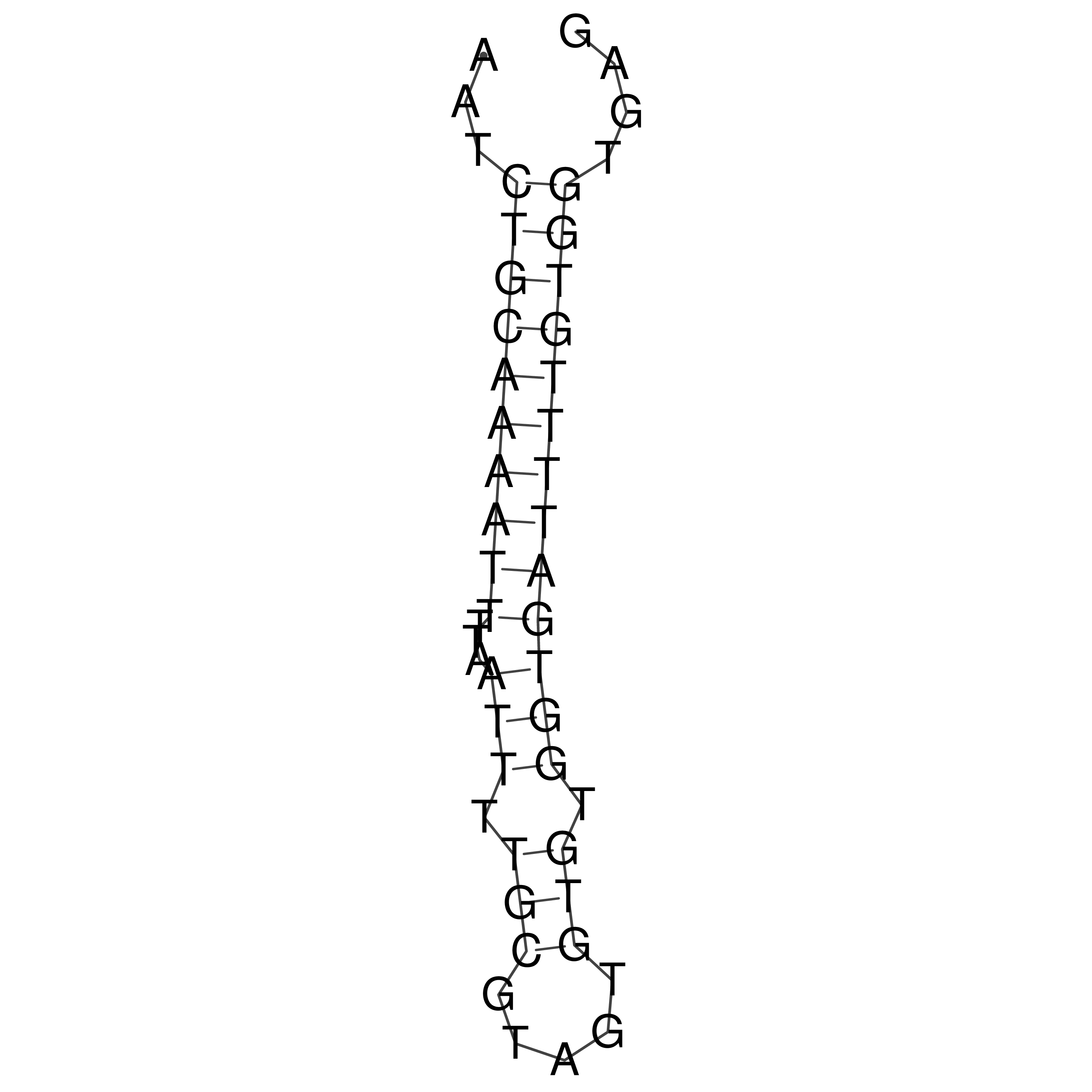
Relaxase
| ID | 3212 | GenBank | WP_181466065 |
| Name | traI_HV185_RS28205_RHBSTW-00405|unnamed |
UniProt ID | _ |
| Length | 1752 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1752 a.a. Molecular weight: 189737.34 Da Isoelectric Point: 5.9228
>WP_181466065.1 conjugative transfer relaxase/helicase TraI [Klebsiella pneumoniae]
MLSFSQVKSAGSAGNYYTEKDNYYVTGSMEERWQGKGAETLELEGKVDKQVFTALLQGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALEQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQEPQIHTHSVVINATQNGDKWQTLGSDTVGKTGFSENILANRIAFGKIYQSTLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKETIDPAEKMVEWMNT
LKETGFDIRGYREAADARAKELASAPAAPVKTDGPDIGDVVTNAIAGLSDRKVQFTYADLLARTVGQLEA
KPGVFERARGGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVTVTPDATVVRLAPF
SDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQNGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGRQTTADIISEPDKGARYNRLAQEFAVSVREGQESVAQISGTREQGVLNGIIRDTLKNEGALGTKEM
AITALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLRDKEGERLDLKVSAVD
SQWSLFRADKLPVANGERLSVLGKIPETRLKGGESITVLNAEAGQLTVQRPGQKTTQALSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDATRTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIATQKAGLHTPAEQVIHLSIPLLESEALNFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGQGNDLLISRQTWDAEKSILTKVLEGKDAVPPLMDSVPGSLMADLTSGQRAATRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQAPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQK
ALSEGEILPAGQPATLYEALVKDYADRTPEAQLQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKAVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAITA
HGAQGASEPYAIALEGVTGGRKQMASFESAYVGLSRMKLHVQVYTDSREGWVNAIQASPSKATAHDILAP
RNERAVKTADLLFGRAQPLNETAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKNAGIW
LSPLTDKDGRLEAIGGDGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMGRQTEQATREVSGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVVSESLQRDRLQQMERDMVRDLSREKTLG
GD
MLSFSQVKSAGSAGNYYTEKDNYYVTGSMEERWQGKGAETLELEGKVDKQVFTALLQGKLPDGSDLTRMQ
DGVNKHRPGYDLTFSAPKSVSMLAMLGGDKRLIDAHNRAVTVALEQVESLASTRVQKDGVSETVLTGNLI
IARFNHDTSRAQEPQIHTHSVVINATQNGDKWQTLGSDTVGKTGFSENILANRIAFGKIYQSTLRADVES
MGYKTVDAGKHGMWEMEGVPVESFSTRSQELREAAGPDASLKSRDVAALDTRKSKETIDPAEKMVEWMNT
LKETGFDIRGYREAADARAKELASAPAAPVKTDGPDIGDVVTNAIAGLSDRKVQFTYADLLARTVGQLEA
KPGVFERARGGIDAAIEREQLIPLDREKGLFTSNIHVLDELSVKALSQEVQRQNHVTVTPDATVVRLAPF
SDAVSVLAQDRPAMGIVSGQGGAAGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVTG
KSALQNGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKESGVNTYR
WQGGRQTTADIISEPDKGARYNRLAQEFAVSVREGQESVAQISGTREQGVLNGIIRDTLKNEGALGTKEM
AITALTPVWLDSKSRGVRDYYREGMVMERWDPETRKHDRFVIDRVTASSNMLTLRDKEGERLDLKVSAVD
SQWSLFRADKLPVANGERLSVLGKIPETRLKGGESITVLNAEAGQLTVQRPGQKTTQALSVGSSPFDGIK
VGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHIRLYSAQDATRTTEKLARHTAFSVVSEQ
LKARSGETDLDAAIATQKAGLHTPAEQVIHLSIPLLESEALNFTRPQLLATALETGGGKVPMTEIDATIQ
AQIKAGSLLNVPVAPGQGNDLLISRQTWDAEKSILTKVLEGKDAVPPLMDSVPGSLMADLTSGQRAATRM
ILETTDRFTVVQGYAGVGKTTQFRAVTGAISLLPEATRPRVIGLGPTHRAVGEMQSAGVDAQTTASFLHD
TQLLQRNGQAPDFSNTLFLLDESSMVGLSDTAKALSLIAAGGGRAVLSGDTDQLQSIAPGQPFRLMQQRS
AADVAIMKEIVRQVPELRPAVYSLIDRDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTPAQEKAIQK
ALSEGEILPAGQPATLYEALVKDYADRTPEAQLQTLIITHLNDDRRVLNSLIHDARRDNGEVGREEVTLP
VLVTSNIRDGELRRLSTWTAHKGAVALVDNVYHRIGSVDKENQLITLMDGEGKERLISPREALAEGVTLY
REQEITVSQGDKMRFSKSDTERGYVANSVWEVKAVSGDSVTLSDGKTTRTLNPKADEAQRHIDLAYAITA
HGAQGASEPYAIALEGVTGGRKQMASFESAYVGLSRMKLHVQVYTDSREGWVNAIQASPSKATAHDILAP
RNERAVKTADLLFGRAQPLNETAAGRAALQQSGLARGQSPGKFISPGKKYPQPHVALPAFDKNGKNAGIW
LSPLTDKDGRLEAIGGDGRVMGNEEARFVALQNSRNGESLLAGNMGEGVRIARDNPDSGVVVRLAGDDRP
WNPEAITGGRIWADPLPNPPINDTGTDIPLPPEVLAQRAAEEAQRREMGRQTEQATREVSGEEKKAGEPG
ERIKEVIGDVIRGLERDRPGEEKTVLPDDPQTRRQEAAVQQVVSESLQRDRLQQMERDMVRDLSREKTLG
GD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1406 | GenBank | WP_020805752 |
| Name | WP_020805752_RHBSTW-00405|unnamed |
UniProt ID | A0A377TIM3 |
| Length | 130 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 130 a.a. Molecular weight: 14772.81 Da Isoelectric Point: 4.5715
>WP_020805752.1 MULTISPECIES: conjugal transfer relaxosome DNA-binding protein TraM [Gammaproteobacteria]
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
MAKIQVYVNDNVAEKINAIAVQRRAEGAKEKDISYSSIASMLLELGLRVYEAQMERKESGFNQMAFNRAL
LESMVKTQFTVNKVLGIECLSPHVNGNPKWEWSGLIENIRDDVSAVMEKFFPNESEIEDE
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | A0A377TIM3 |
T4CP
| ID | 3126 | GenBank | WP_064343540 |
| Name | traD_HV185_RS28200_RHBSTW-00405|unnamed |
UniProt ID | _ |
| Length | 764 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 764 a.a. Molecular weight: 85378.44 Da Isoelectric Point: 4.9296
>WP_064343540.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTVCVVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIQRRLDARVDGRLSALLEAREAEGSLARALFTPDEPVSESAEEKAGVQPEQTKPAEDS
VLPVPVKTSSKAAEGAGKTPDVPLSGAPVTRVKTVPLIRPEPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLAGEQTQRDMQRREEVNINHSRSEQDDIEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLAPMR
DIIRSQPVYTISYYGKSLEYNAEQILADKYTIWCGEQLWTAFVFAAIVSLTVCVVTFFVASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGMASDIKIGDLPILLNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDKDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDQPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKITEGFIQRRLDARVDGRLSALLEAREAEGSLARALFTPDEPVSESAEEKAGVQPEQTKPAEDS
VLPVPVKTSSKAAEGAGKTPDVPLSGAPVTRVKTVPLIRPEPAASVTGTAAAASTAAVSATAGGTEQELA
QQPVEADQDMLPAGMNEDGVIEDMQAYDAWLAGEQTQRDMQRREEVNINHSRSEQDDIEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 71216..99198
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HV185_RS28000 (HV185_28010) | 66408..67283 | + | 876 | WP_060415463 | DUF4942 domain-containing protein | - |
| HV185_RS29565 | 67665..67773 | - | 109 | Protein_85 | hypothetical protein | - |
| HV185_RS28005 (HV185_28015) | 68061..68399 | + | 339 | WP_020804855 | hypothetical protein | - |
| HV185_RS28010 (HV185_28020) | 68456..68803 | + | 348 | WP_064182675 | hypothetical protein | - |
| HV185_RS28015 (HV185_28025) | 68892..69122 | + | 231 | WP_060415465 | hypothetical protein | - |
| HV185_RS28020 (HV185_28030) | 69169..70002 | + | 834 | WP_060415466 | N-6 DNA methylase | - |
| HV185_RS28025 (HV185_28035) | 70288..70656 | - | 369 | WP_080848162 | hypothetical protein | - |
| HV185_RS28030 (HV185_28040) | 70651..71193 | + | 543 | WP_060415467 | antirestriction protein | - |
| HV185_RS28035 (HV185_28045) | 71216..71638 | - | 423 | WP_073972322 | transglycosylase SLT domain-containing protein | virB1 |
| HV185_RS28040 (HV185_28050) | 72142..72534 | + | 393 | WP_020805752 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| HV185_RS29570 | 72772..73473 | + | 702 | WP_223174962 | hypothetical protein | - |
| HV185_RS28050 (HV185_28060) | 73559..73759 | + | 201 | WP_048289787 | TraY domain-containing protein | - |
| HV185_RS28055 (HV185_28065) | 73828..74196 | + | 369 | WP_064343525 | type IV conjugative transfer system pilin TraA | - |
| HV185_RS28060 (HV185_28070) | 74210..74515 | + | 306 | WP_064343526 | type IV conjugative transfer system protein TraL | traL |
| HV185_RS28065 (HV185_28075) | 74531..75097 | + | 567 | WP_064359954 | type IV conjugative transfer system protein TraE | traE |
| HV185_RS28070 (HV185_28080) | 75084..75818 | + | 735 | WP_064343528 | type-F conjugative transfer system secretin TraK | traK |
| HV185_RS28075 (HV185_28085) | 75818..77239 | + | 1422 | WP_053390199 | F-type conjugal transfer pilus assembly protein TraB | traB |
| HV185_RS28080 (HV185_28090) | 77232..77666 | + | 435 | WP_181466063 | conjugal transfer protein TraP | - |
| HV185_RS28085 (HV185_28095) | 77700..78284 | + | 585 | WP_064343530 | type IV conjugative transfer system lipoprotein TraV | traV |
| HV185_RS28090 (HV185_28100) | 78897..79085 | + | 189 | WP_053390286 | hypothetical protein | - |
| HV185_RS29575 | 79169..79570 | + | 402 | WP_064343532 | hypothetical protein | - |
| HV185_RS28100 (HV185_28110) | 79567..79866 | + | 300 | WP_181466064 | hypothetical protein | - |
| HV185_RS28105 (HV185_28115) | 79954..80337 | + | 384 | WP_042946290 | hypothetical protein | - |
| HV185_RS28110 (HV185_28120) | 80430..83069 | + | 2640 | WP_064343533 | type IV secretion system protein TraC | virb4 |
| HV185_RS28115 (HV185_28125) | 83069..83461 | + | 393 | WP_064343534 | type-F conjugative transfer system protein TrbI | - |
| HV185_RS28120 (HV185_28130) | 83458..84084 | + | 627 | WP_064343535 | type-F conjugative transfer system protein TraW | traW |
| HV185_RS28125 (HV185_28135) | 84128..85087 | + | 960 | WP_229295972 | conjugal transfer pilus assembly protein TraU | traU |
| HV185_RS28130 (HV185_28140) | 85100..85747 | + | 648 | WP_064343536 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| HV185_RS28135 (HV185_28145) | 85744..86127 | + | 384 | WP_053390297 | hypothetical protein | - |
| HV185_RS28140 (HV185_28150) | 86124..88079 | + | 1956 | WP_064359956 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| HV185_RS28145 (HV185_28155) | 88111..88371 | + | 261 | WP_053390216 | hypothetical protein | - |
| HV185_RS28150 (HV185_28160) | 88364..88975 | + | 612 | WP_053390217 | hypothetical protein | - |
| HV185_RS28155 (HV185_28165) | 88965..89207 | + | 243 | WP_053390218 | conjugal transfer protein TrbE | - |
| HV185_RS28160 (HV185_28170) | 89253..89579 | + | 327 | WP_053390219 | hypothetical protein | - |
| HV185_RS28165 (HV185_28175) | 89600..90352 | + | 753 | WP_053390220 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| HV185_RS28170 (HV185_28180) | 90363..90599 | + | 237 | WP_053390221 | type-F conjugative transfer system pilin chaperone TraQ | - |
| HV185_RS28175 (HV185_28185) | 90574..91134 | + | 561 | WP_039698500 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| HV185_RS28180 (HV185_28190) | 91121..92491 | + | 1371 | WP_049078362 | conjugal transfer pilus assembly protein TraH | traH |
| HV185_RS28185 (HV185_28195) | 92491..95334 | + | 2844 | WP_049078361 | conjugal transfer mating-pair stabilization protein TraG | traG |
| HV185_RS28190 (HV185_28200) | 95340..95855 | + | 516 | WP_049078360 | conjugal transfer protein TraS | - |
| HV185_RS28195 (HV185_28205) | 95918..96664 | + | 747 | WP_049078359 | hypothetical protein | - |
| HV185_RS28200 (HV185_28210) | 96904..99198 | + | 2295 | WP_064343540 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 4847 | GenBank | NZ_CP055520 |
| Plasmid name | RHBSTW-00405|unnamed | Incompatibility group | ColRNAI |
| Plasmid size | 112388 bp | Coordinate of oriT [Strand] | 71783..71831 [-] |
| Host baterium | Klebsiella pneumoniae strain RHBSTW-00405 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |