Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104374 |
| Name | oriT_pCTYH.Ch1_2 |
| Organism | Ensifer adhaerens strain CTYH.Ch1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP118612 (554944..554975 [+], 32 nt) |
| oriT length | 32 nt |
| IRs (inverted repeats) | 17..22, 27..32 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 32 nt
>oriT_pCTYH.Ch1_2
GAAGGGCGCAATTATACGTCGCTGTTGCGACG
GAAGGGCGCAATTATACGTCGCTGTTGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file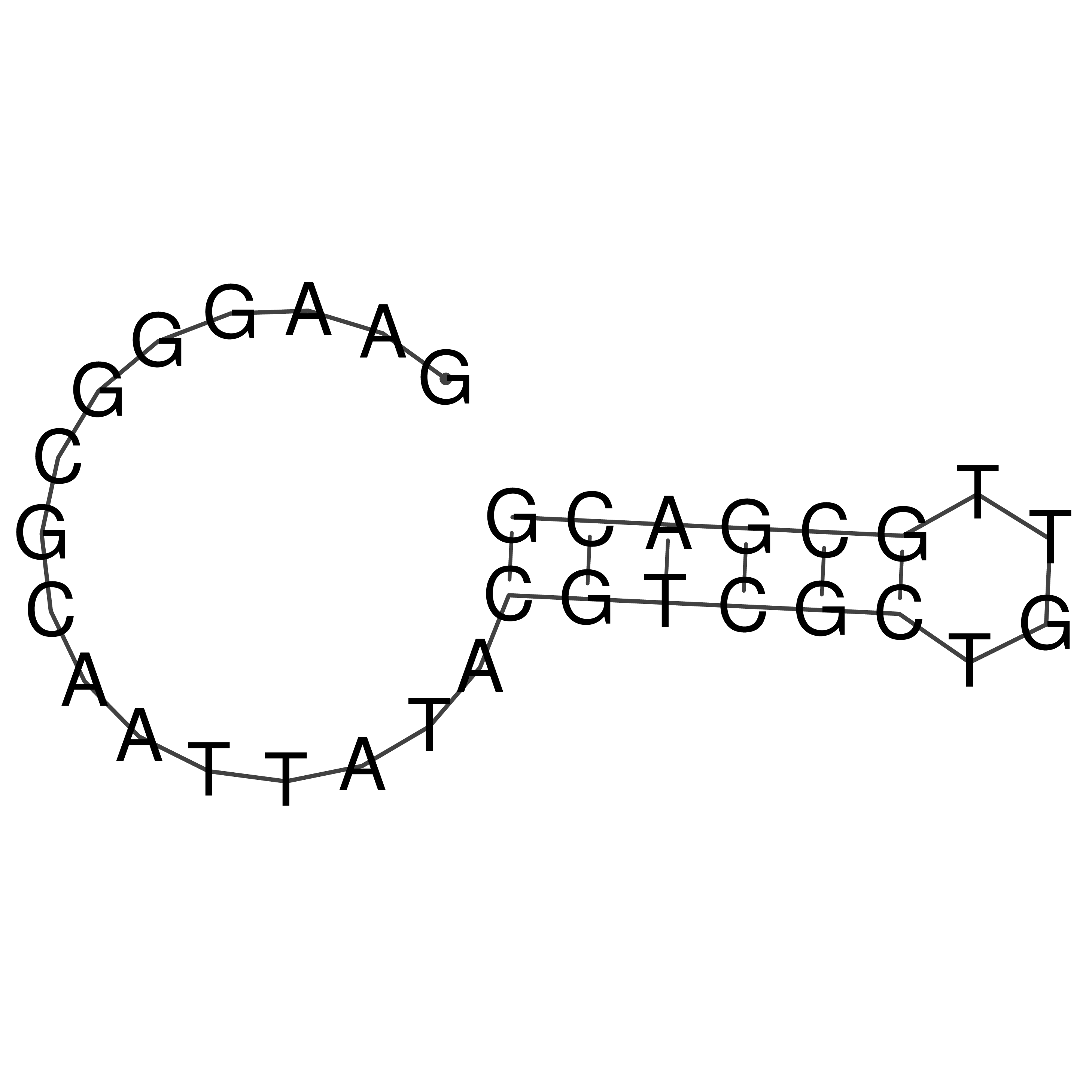
Relaxase
| ID | 3196 | GenBank | WP_275027580 |
| Name | traA_PWG15_RS35560_pCTYH.Ch1_2 |
UniProt ID | _ |
| Length | 1101 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1101 a.a. Molecular weight: 121242.64 Da Isoelectric Point: 10.0236
>WP_275027580.1 Ti-type conjugative transfer relaxase TraA [Ensifer adhaerens]
MAVPHFSVNIVARGAGRSAVMSAAYRHCAKMDFEREARGVDYRAKQGLLHEEFVVPADAPAWLSSMIADR
SVSGASEAFWNRVEEFEKRADAQLAKDLTIALPIELTPEQNIGLVREFVERQILSKGMVADWVYHDAPGN
PHVHLMATLRPLTDEGFGAKKVALLAPDGRPLRNDMGKIVYELWAGGVDDFNALRDGWFACQNRHLALAG
LDIRVDGRSYESQGITVAPTIHLGVGAKAIERKSTASQMPASLERLELQEALRQANARRIERRPQIVLDM
ITREKSVFDERDVAKILHRYIDDADLFGSLMLRVLRDPSALRLTAEGTDLGTGLYSPARYTTRELIRLEA
EMVNRALWLARRTSHGVRGALLEATFSKNRGLSDEQQAAIEHLTTGAALAAVVGRAGAGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWERRWAQGTDCLDEKTVFVLDEAGMVSSRQMAFFVE
AVVRSGAKLVLVGDPEQLQPIEAGAAFRAIGDRVGYAELQTIYRQRAPWMRSASLDLARGKIGEAIDAYR
ANGKIVGAELKSEAIERLITDWDREHDLTKTSLMLAHLRRDVRLLNSLARARLVERGVVGEGAAFRTADG
NRNFSAGDQIVFLKNDGILGVKNGMLGRVVEAGPGRFIASIGEGTHRRDVTVEQRLYNNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDELAVYYGRRSFAKSGGLVPILSRRNAKETTLDYEARPLY
RQALRFAEARGLHLMSVARTVVRDRLHWIVGQKQKLAMLGARLVDLASSIGLARGKLERAAPAIGAARPI
VSAITVFAKSVEQAVADRLAADLNLRERWQDVSMRFQLVYADPQVAFAAVKVDAMLKDTSLAATVLQELA
AEPQHFGALSGRTGMFASRSDKKDRDAALLNVPALARNLADYVRVRAETERRCLAEENAVRSKLAVEIPA
LSPVTRQALEQLREGLERDDQARHEPSPLPPSIRQELERFSAAVSQRFGERTFPQIAAKAPDGEVFAAVT
AGMAPQQKNEVRLAWDAMRTIQLLSARDRVAAGLKLSETQRQSQGKELRLK
MAVPHFSVNIVARGAGRSAVMSAAYRHCAKMDFEREARGVDYRAKQGLLHEEFVVPADAPAWLSSMIADR
SVSGASEAFWNRVEEFEKRADAQLAKDLTIALPIELTPEQNIGLVREFVERQILSKGMVADWVYHDAPGN
PHVHLMATLRPLTDEGFGAKKVALLAPDGRPLRNDMGKIVYELWAGGVDDFNALRDGWFACQNRHLALAG
LDIRVDGRSYESQGITVAPTIHLGVGAKAIERKSTASQMPASLERLELQEALRQANARRIERRPQIVLDM
ITREKSVFDERDVAKILHRYIDDADLFGSLMLRVLRDPSALRLTAEGTDLGTGLYSPARYTTRELIRLEA
EMVNRALWLARRTSHGVRGALLEATFSKNRGLSDEQQAAIEHLTTGAALAAVVGRAGAGKTTMMKAAREA
WQAAGYRVVGGALAGKAAEGLEKEAGIASRTLSSWERRWAQGTDCLDEKTVFVLDEAGMVSSRQMAFFVE
AVVRSGAKLVLVGDPEQLQPIEAGAAFRAIGDRVGYAELQTIYRQRAPWMRSASLDLARGKIGEAIDAYR
ANGKIVGAELKSEAIERLITDWDREHDLTKTSLMLAHLRRDVRLLNSLARARLVERGVVGEGAAFRTADG
NRNFSAGDQIVFLKNDGILGVKNGMLGRVVEAGPGRFIASIGEGTHRRDVTVEQRLYNNVDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHRDELAVYYGRRSFAKSGGLVPILSRRNAKETTLDYEARPLY
RQALRFAEARGLHLMSVARTVVRDRLHWIVGQKQKLAMLGARLVDLASSIGLARGKLERAAPAIGAARPI
VSAITVFAKSVEQAVADRLAADLNLRERWQDVSMRFQLVYADPQVAFAAVKVDAMLKDTSLAATVLQELA
AEPQHFGALSGRTGMFASRSDKKDRDAALLNVPALARNLADYVRVRAETERRCLAEENAVRSKLAVEIPA
LSPVTRQALEQLREGLERDDQARHEPSPLPPSIRQELERFSAAVSQRFGERTFPQIAAKAPDGEVFAAVT
AGMAPQQKNEVRLAWDAMRTIQLLSARDRVAAGLKLSETQRQSQGKELRLK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3109 | GenBank | WP_275027575 |
| Name | traG_PWG15_RS35545_pCTYH.Ch1_2 |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 68606.28 Da Isoelectric Point: 9.0825
>WP_275027575.1 Ti-type conjugative transfer system protein TraG [Ensifer adhaerens]
MGVSRIALAASPLALMALALVTMEGAGDWLSSFAGREAGRLALIRAGVAIPSVSAAVVGAIFLFAAAGSA
NIRFAGWGAFGGGLTAAAVALIDGFVDTSLAQSWQFHTLDFEPAAVVGAVAALLSSGFALRVAMIGNAAF
PKSAPKRVRGKRAIYGEAEWMKLSCAKRLFPEVAGIVIGEAYRVDEDGVAAIPFRADKPETWGAGGRSPL
LCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGSTLVVLDPSNEVAPMVIDHRKGAGRDVRILDPKKP
DAGFNALDWIGRYGRSKEEDIAAVASWIMSDSGRAIGVRDDFFRASALQLLTALIADVCLSGHTRKEDQT
LRQVRRNLAEPEPKLRERLQAIHANSSSGFVRENVAAFVNMTPETFSGVYANAVKETHWLSYENYAGLVS
SKSFSTEQLLAGSLDVFINIDLKTLETHSGLARVIIGSFVNAIYNRNRSGDDLALFLLDEVARLGYLRIL
ETARDTGRKYGISLTLIFQSIGQMRETFGGRDATSKWFESASWVSFAAINDPETAEYVSRRCGMTTVEVD
QVSRSFQSRGSSRTRSKQLAARPLIQPDEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMQGCVKGSRF
HAPSEPV
MGVSRIALAASPLALMALALVTMEGAGDWLSSFAGREAGRLALIRAGVAIPSVSAAVVGAIFLFAAAGSA
NIRFAGWGAFGGGLTAAAVALIDGFVDTSLAQSWQFHTLDFEPAAVVGAVAALLSSGFALRVAMIGNAAF
PKSAPKRVRGKRAIYGEAEWMKLSCAKRLFPEVAGIVIGEAYRVDEDGVAAIPFRADKPETWGAGGRSPL
LCFDGSFGSSHGIVFAGSGGFKTTSVTVPTALKWGSTLVVLDPSNEVAPMVIDHRKGAGRDVRILDPKKP
DAGFNALDWIGRYGRSKEEDIAAVASWIMSDSGRAIGVRDDFFRASALQLLTALIADVCLSGHTRKEDQT
LRQVRRNLAEPEPKLRERLQAIHANSSSGFVRENVAAFVNMTPETFSGVYANAVKETHWLSYENYAGLVS
SKSFSTEQLLAGSLDVFINIDLKTLETHSGLARVIIGSFVNAIYNRNRSGDDLALFLLDEVARLGYLRIL
ETARDTGRKYGISLTLIFQSIGQMRETFGGRDATSKWFESASWVSFAAINDPETAEYVSRRCGMTTVEVD
QVSRSFQSRGSSRTRSKQLAARPLIQPDEVLRMRADEQIVFTAGNAPLRCGRAIWFRREDMQGCVKGSRF
HAPSEPV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3110 | GenBank | WP_275028018 |
| Name | t4cp2_PWG15_RS35850_pCTYH.Ch1_2 |
UniProt ID | _ |
| Length | 816 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 816 a.a. Molecular weight: 91314.23 Da Isoelectric Point: 5.6099
>WP_275028018.1 conjugal transfer protein TrbE [Ensifer adhaerens]
MVNLRRFRSPDPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESATDLERNEISRQINAVLARLG
SGWMIQVEAVRVEATAYPSAERSHFPDPVTRAIEAERRAHFEREGTHFESRHALILTQRPIEQRRSGLGK
YVYADPESRKRGFADTALDLFRTRVREVEQYLANWISLRRMATRELNPADDGRKERFDELFQFIRFCISG
ENHPIRLPDIPFYLDWLVTAEFQHGLTPLVDNRHLGVVAIDGLPSESWPGILNGLDRLPMAYRWSSRFIF
LDAQEARERLERTRKKWQQKVRPFFDQLFQTEGRSIDQDAIAMVAEVEDAIAQASSELVAYGYYTPVVIV
FGADEPEVRDKCELVRQAVQVEGFGARIETLNATEAYLGSLPGNWYCNVREPLINTRNLADLIPLNTVWA
GNPFAPCPYLPPNSPPLLQVASGSTPFRFNLHVDDIGHTLVFGPTGSGKSTLLALIAAQFRRYVGAQVFI
FDKGRSMLPLTLAVGGGHYEIGADGEAPGLCFCPLLDLEGEGDRAWACQWVETLAELQGASIGPEERNAI
SRQITLMATASGRSLSDFVSGVQLKAIKDALQHYTIDGPLGALIDAEIDTLSLGAFEAFEIEHLMVLGER
SLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPLFRERIREWLKVLRKANCAVVLATQSISDAERSG
IIDVLRESCPTKICLPNAAARQPGAREFYERMGFNDRQIEIVAASTPKRDYYVTSPEGRRLFDMALGPLA
LAFVGAAGKDDLKRIRALHLEHGRSWPVEWLKLRGIPDAETLLADS
MVNLRRFRSPDPSFADLVPYAGLVDNGIILLKDGSLMAGWYFAGPDSESATDLERNEISRQINAVLARLG
SGWMIQVEAVRVEATAYPSAERSHFPDPVTRAIEAERRAHFEREGTHFESRHALILTQRPIEQRRSGLGK
YVYADPESRKRGFADTALDLFRTRVREVEQYLANWISLRRMATRELNPADDGRKERFDELFQFIRFCISG
ENHPIRLPDIPFYLDWLVTAEFQHGLTPLVDNRHLGVVAIDGLPSESWPGILNGLDRLPMAYRWSSRFIF
LDAQEARERLERTRKKWQQKVRPFFDQLFQTEGRSIDQDAIAMVAEVEDAIAQASSELVAYGYYTPVVIV
FGADEPEVRDKCELVRQAVQVEGFGARIETLNATEAYLGSLPGNWYCNVREPLINTRNLADLIPLNTVWA
GNPFAPCPYLPPNSPPLLQVASGSTPFRFNLHVDDIGHTLVFGPTGSGKSTLLALIAAQFRRYVGAQVFI
FDKGRSMLPLTLAVGGGHYEIGADGEAPGLCFCPLLDLEGEGDRAWACQWVETLAELQGASIGPEERNAI
SRQITLMATASGRSLSDFVSGVQLKAIKDALQHYTIDGPLGALIDAEIDTLSLGAFEAFEIEHLMVLGER
SLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPLFRERIREWLKVLRKANCAVVLATQSISDAERSG
IIDVLRESCPTKICLPNAAARQPGAREFYERMGFNDRQIEIVAASTPKRDYYVTSPEGRRLFDMALGPLA
LAFVGAAGKDDLKRIRALHLEHGRSWPVEWLKLRGIPDAETLLADS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 613511..622792
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PWG15_RS35800 (PWG15_35795) | 609028..610374 | - | 1347 | WP_275027631 | amidohydrolase | - |
| PWG15_RS35805 (PWG15_35800) | 610391..611398 | - | 1008 | WP_275028015 | response regulator transcription factor | - |
| PWG15_RS35810 (PWG15_35805) | 611895..612626 | + | 732 | WP_275027632 | winged helix-turn-helix domain-containing protein | - |
| PWG15_RS35815 (PWG15_35810) | 612780..613496 | + | 717 | WP_275027633 | autoinducer binding domain-containing protein | - |
| PWG15_RS35820 (PWG15_35815) | 613511..614791 | - | 1281 | WP_275027634 | IncP-type conjugal transfer protein TrbI | virB10 |
| PWG15_RS35825 (PWG15_35820) | 614801..615241 | - | 441 | WP_275027635 | conjugal transfer protein TrbH | - |
| PWG15_RS35830 (PWG15_35825) | 615246..616034 | - | 789 | WP_275028016 | P-type conjugative transfer protein TrbG | virB9 |
| PWG15_RS35835 (PWG15_35830) | 616076..616738 | - | 663 | WP_275027636 | conjugal transfer protein TrbF | virB8 |
| PWG15_RS35840 (PWG15_35835) | 616759..617889 | - | 1131 | WP_275027637 | P-type conjugative transfer protein TrbL | virB6 |
| PWG15_RS35845 (PWG15_35840) | 617943..618737 | - | 795 | WP_275027638 | P-type conjugative transfer protein TrbJ | virB5 |
| PWG15_RS35850 (PWG15_35845) | 618709..621159 | - | 2451 | WP_275028018 | conjugal transfer protein TrbE | virb4 |
| PWG15_RS35855 (PWG15_35850) | 621168..621467 | - | 300 | WP_275027639 | conjugal transfer protein TrbD | virB3 |
| PWG15_RS35860 (PWG15_35855) | 621460..621837 | - | 378 | WP_275027640 | TrbC/VirB2 family protein | virB2 |
| PWG15_RS35865 (PWG15_35860) | 621827..622792 | - | 966 | WP_275027641 | P-type conjugative transfer ATPase TrbB | virB11 |
| PWG15_RS35870 (PWG15_35865) | 622789..623424 | - | 636 | WP_275027642 | acyl-homoserine-lactone synthase | - |
Host bacterium
| ID | 4813 | GenBank | NZ_CP118612 |
| Plasmid name | pCTYH.Ch1_2 | Incompatibility group | - |
| Plasmid size | 623840 bp | Coordinate of oriT [Strand] | 554944..554975 [+] |
| Host baterium | Ensifer adhaerens strain CTYH.Ch1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | hdt |
| Symbiosis gene | fixA, fixB |
| Anti-CRISPR | - |