Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104348 |
| Name | oriT_2020CK-00216|unnamed1 |
| Organism | Enterobacter kobei strain 2020CK-00216 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP118561 (82602..82704 [-], 103 nt) |
| oriT length | 103 nt |
| IRs (inverted repeats) | 89..94, 96..101 (AAATAA..TTATTT) 28..33, 40..45 (GCAAAA..TTTTGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 103 nt
>oriT_2020CK-00216|unnamed1
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
CTCACCACCAAAAGCTCCACACCCCACGCAAAATTAAAGTTTTGCTGATTGGGTATTCAAATCATGCAGTTAGTGAAAAAGTAATGTGAAATAATTTATTTTA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file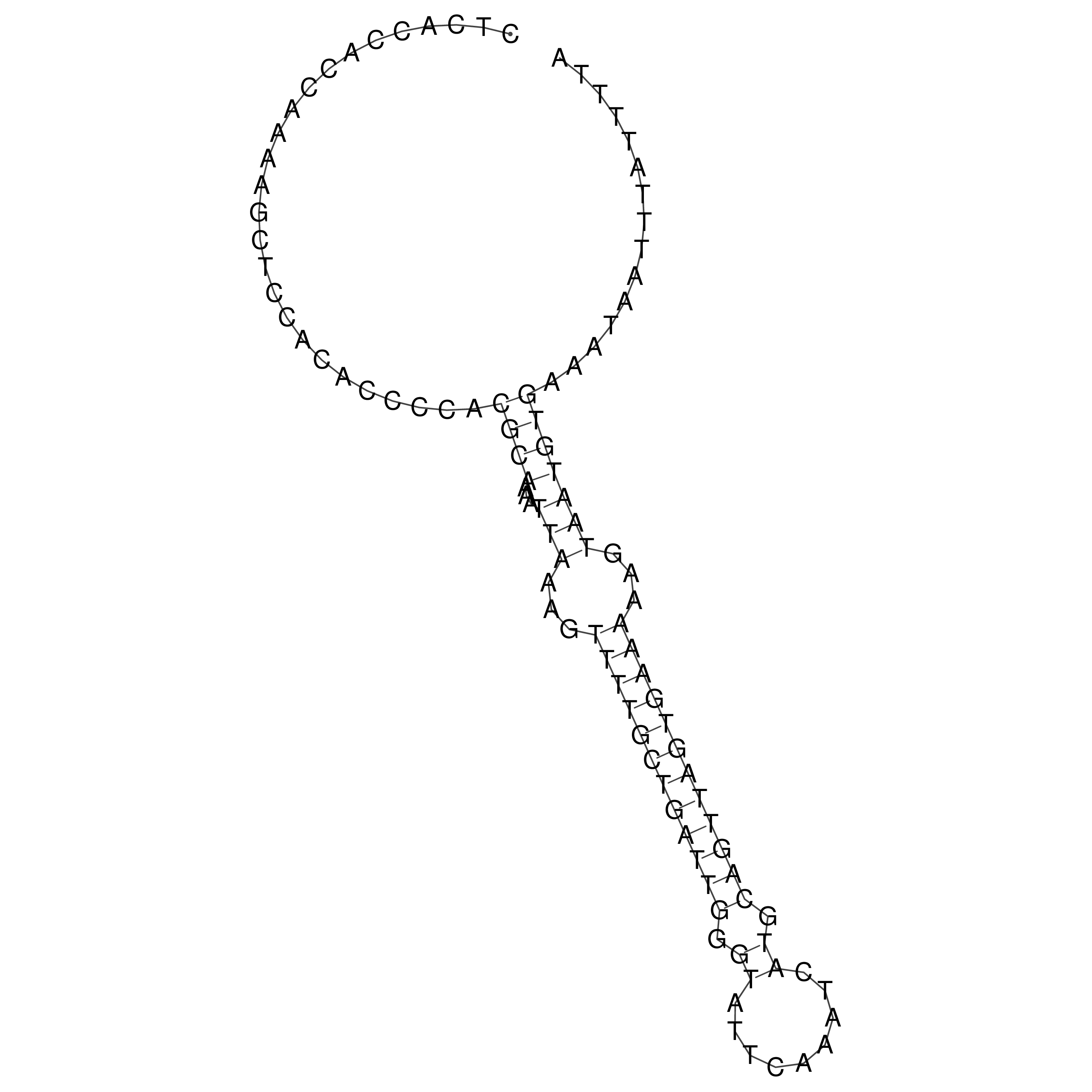
Relaxase
| ID | 3181 | GenBank | WP_023307204 |
| Name | traI_PXH70_RS23065_2020CK-00216|unnamed1 |
UniProt ID | A0A1Q1PS85 |
| Length | 1746 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1746 a.a. Molecular weight: 189264.44 Da Isoelectric Point: 6.1121
>WP_023307204.1 MULTISPECIES: conjugative transfer relaxase/helicase TraI [Enterobacterales]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMGERWAGKGAEALGLSGGVDQKVFTKVLEGRLPDGSDLSRTQ
DGANKHRPGYDFTFSAPKSVSVMAMLGGDKRLIEAHNRAVETAIKQIEAMASTRVMTEGRSETQLTGNLV
MALFNHDTSRDQEPQLHTHAVVANATQHGDEWRTLSSDKVGKTGFIENIYANQIAFGRLYRAALKDDVAA
MGYETETVGKHGMWELKGVPTEPYSSRSRTISEAVGDDASLKSRDVAALDTRQSKQKVDPEQRMAEWMHT
LKETGFDIKAYREAADLRVVQGNIPATTPEAIDINSSVGQAIAMLSDRRARFTYSELLATTLGQLPARSG
MVEMARDGIDAAIKNEQLIPLDKEKGLFTSNIHVLDELSVSAMTRDLQRTGQADIFPDKSVPRSRSYSDA
VSVLAQDRPPVAIISGQGGAAGQRERVAELTMMAREQGRDVQIIAADRRSRSALMQDERLSGEHITDRRG
LTEGMAFTPGSTLIVDQGEKLTLKETLTLLDGALRHNVQLLIADSGQRTGTGSALTVMKEAGVSTLAWQG
GEKTQVSVISEPDRRQRYDRLAADFARSVRAGEHSVAQVTGPREQAVLAGVVREALKAEKVLGEREVSIT
TLEPVWLDSRHQQVRDHYREGMVMERWNAEERSRERFVIDRVTGRNNSLTLRNAQGETRVTRLSELNSSW
SLYRTGTLQVAEGDRLAVLGQAQGTRLRGGDTVTVKALGEAGLVVSLPGRKADVVLPAGDSPFTAAKVVQ
GWVESPGRSVSDSATVFASLSQRELDNTTLNKLALSGRAVRLYSAQTAQKTTEKLSRQSAWSVVSEQIKD
AAGHDRLDDALAHQKAGLHTPEQQAIHLAIPVLEGNGLAFTKPQLMAAAKEFEQDSLSLQAIEKEADRQV
RSGDLLSVPVSPGNGLQLLVSRQSYNAEKSIIRHVLEGKEAVTPLMDKVPDAQLAGLTDGQRNATRMILE
SPDRFTLVQGYAGVGKTTQFRAVMSAIGTLPEREQPRVIGVAPTHRAVSEMRDAGVPETQTLAAFIHDTQ
QQLRGGERTDFSNVLFLVDESSMVGNADMAKAYSLIAGGGGRAVSSGDTDQLQSIAPGQPFRLLQKRSAI
DVAVMQEIVRQTPELKPAVYSLISRDIGSALTTIESVAPAQVPRRADAWQPDSSVMEFSREREEAIAKAV
AAGDLMPGGQPATLLEAIVKDYTGRTSEAQAQTIVITALNADRRQVNAMIHDARQGAGEVGEKEVTLPVL
TPANIRDGELRRMATWEASRDSLVLLDSTYYSIEALDGEAHLVMLKDAQGNTRSLSPAQAATEGVTLYRQ
DTITVSEGDRMRFSKSDNERGFVANSVWSVSEIKGDSVTLSDGKQTRTLTPSAEQAEQHIDLAYAVTVEG
SQGASEPFSISLQGTDGGRKRMVSFESAYVALSRMKQHAQVYTDNRDKWVAAMEKSQAKSTAHDILEPRG
DRAVANAARLTATAKALGEVPAGRAALRQAGLQPEGSMAKYISPGRKYPQPHVALPAFDRNGRKAGVWLS
ALTSGDGQLKGLAGEGRVMGSGDAAFAGLQASRNGESLLARDMEEGVRLARENPQSGVVVRLEGEARPWN
PGAITGGRVWADGLPDATGTQPAETVPPEVLAQQAREAEVQRELDKRAEEAVREMARSGDRPAGGADVAV
RDVVRDMDRIKETPASPLTLPDAPEERRRDEAVSQVVRESVQRDRLQQMERETVRDLEREKTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3096 | GenBank | WP_022652256 |
| Name | traD_PXH70_RS23070_2020CK-00216|unnamed1 |
UniProt ID | _ |
| Length | 736 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 736 a.a. Molecular weight: 82566.18 Da Isoelectric Point: 5.1609
>WP_022652256.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacterales]
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAW
QAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
MSFNAKDMTQGGQIASMRIRMFSQIANIIVYCLVILFGIIVAITLSVKLTWQTFVNGMVYYWCQTLAPMR
DIFKSQPVYTIRYYGHELKYNAQQILQDKYIQWCADQVWTAFILAAIIAAVVCVITFFVTTWILGHQGKQ
QSEDEITGGRQLTDDPKAVAKMLRRDGLASDIKIGELPLIKDSEIQNFCLHGTVGAGKSEVIRRLMNYAR
QRGDMVIVYDRSCEFVKSYYDPATDKILNPLDARCAAWDLWRECLTQPDFDNVSNTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDKDRSYSKLVDTLLSIKIEKLRSYLKNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIENNGESFTIRDWMRGVREDGKNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVIGIQSYAQLEDIYGQKPAETLFDVLNTRAFFRSPSRNIA
EFAAGEIGEKEILKASEQYSYGVDPVRDGTSVGKDMERVTLVSYSDVQTLPDLSCYVTLPGPYPAVKMAL
KYQQRPKVAPEFIPRTIDGKAEERLNAALAAREHESRSIAMLFEPDADVVAPEGGAENVGQPQQPQQPQQ
PQQPQQPQQPQQPQQLTPAPRAADAASVAGAAGAGGVEPELKTKAEEAEQLPPGISESGEVVDMAAWEAW
QAEQDELSPQERQRREEVNINVHRAPEKDLEPGGYI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 3097 | GenBank | WP_008324134 |
| Name | traC_PXH70_RS23145_2020CK-00216|unnamed1 |
UniProt ID | _ |
| Length | 876 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 876 a.a. Molecular weight: 98841.51 Da Isoelectric Point: 5.7676
>WP_008324134.1 MULTISPECIES: type IV secretion system protein TraC [Enterobacterales]
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
MSNPLDSVTRTVNSLLTALKMPDASSEANDVLGQMRFPQFSRVLPYRDYDAETGLFMNEKTMGFMLEAQP
INGANETIVSSLENLMRTKLPREVPVAVHLVSSHLVGQDIEYGLRELSWTGEKADTFNAITRAYYLRAAE
EGFPLPDHLNLPLTLRNYRVYISCCVPRKRGSKASVMEMENQVKIIRASLQGAQIATRSVDADAFIRIAG
EMINHNPDALYPTERELDPLKDLNYQCVDDTFDLKVKADYMTLGLRGNGRNSTARIMNFQLTKNPELAFL
WNMADNYSNLLNPELSVSCPFILTLVLSVEDQVKAQNEANLKFMDLEKKSKTSYAKFFPGVAQEAKEWGD
LRQRLSSGQSSLVSYFLNVTLFCRDDQETALKVEQDVLNSFRKNGFELISPRFNHMRNFLVNLPFMAGGG
LFKQLRESGVLQRAESFNVANLMPVVADSPLATSGLLAPTYRNQLAFIDLFSRGMGNTNFNMAVCGTSGA
GKTGLIQPLIRSVIDSGGFAVVFDMGDGYKSLCENMGGVYLDGETLKFNPFANVTDDTIDEMAERLRDQL
SVMASPNGNLDEVHEGLLLKAVKATWLTKKNQARIDDVVDYLMMARDSEEYSGSPTIRSRLDEMVVLLTQ
YTRDGVYGSYFNSDEPSLKDEARMVVLELGGLEDRPSLLVAVMFSLILYIENRMYRTPRSLKKLNVIDEG
WRLLDFKNNKVGAFIEKGYRTARRHTGSYITITQNIVDFDSETASSAARAAWGNSSYKIILKQSAKEFAK
YNQLYPDQYSQLQRDMIKGFGSAKDNWFSSFMLSVEHLTSWHRLFVDPLSRAMYSSDGPDFEFLQQRRRE
GLSIHEAVWELAWKKSGPEMAALERWLHEHEGKTIS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 58118..83252
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PXH70_RS23070 (PXH70_23070) | 58118..60328 | - | 2211 | WP_022652256 | type IV conjugative transfer system coupling protein TraD | virb4 |
| PXH70_RS23075 (PXH70_23075) | 60462..61136 | - | 675 | WP_008324909 | tetratricopeptide repeat protein | - |
| PXH70_RS23080 (PXH70_23080) | 61211..61642 | - | 432 | WP_008324118 | entry exclusion protein | - |
| PXH70_RS23085 (PXH70_23085) | 61664..64486 | - | 2823 | WP_008324119 | conjugal transfer mating-pair stabilization protein TraG | traG |
| PXH70_RS23090 (PXH70_23090) | 64486..65856 | - | 1371 | WP_016247527 | conjugal transfer pilus assembly protein TraH | traH |
| PXH70_RS23095 (PXH70_23095) | 65843..66271 | - | 429 | WP_008324122 | hypothetical protein | - |
| PXH70_RS23100 (PXH70_23100) | 66264..66818 | - | 555 | WP_008324125 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| PXH70_RS23105 (PXH70_23105) | 66808..67056 | - | 249 | WP_008324126 | type-F conjugative transfer system pilin chaperone TraQ | - |
| PXH70_RS23110 (PXH70_23110) | 67073..67822 | - | 750 | WP_274850492 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| PXH70_RS23115 (PXH70_23115) | 67853..68041 | - | 189 | WP_008324128 | conjugal transfer protein TrbE | - |
| PXH70_RS23120 (PXH70_23120) | 68073..69926 | - | 1854 | WP_022652255 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| PXH70_RS23125 (PXH70_23125) | 69923..70552 | - | 630 | WP_008324130 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| PXH70_RS23130 (PXH70_23130) | 70569..71555 | - | 987 | WP_032160671 | conjugal transfer pilus assembly protein TraU | traU |
| PXH70_RS23135 (PXH70_23135) | 71558..72205 | - | 648 | WP_008324132 | type-F conjugative transfer system protein TraW | traW |
| PXH70_RS23140 (PXH70_23140) | 72205..72552 | - | 348 | WP_008324133 | type-F conjugative transfer system protein TrbI | - |
| PXH70_RS23145 (PXH70_23145) | 72549..75179 | - | 2631 | WP_008324134 | type IV secretion system protein TraC | virb4 |
| PXH70_RS23150 (PXH70_23150) | 75172..75738 | - | 567 | WP_008324135 | conjugal transfer pilus-stabilizing protein TraP | - |
| PXH70_RS23155 (PXH70_23155) | 75728..76129 | - | 402 | WP_008324137 | hypothetical protein | - |
| PXH70_RS23160 (PXH70_23160) | 76113..76298 | - | 186 | WP_008324139 | hypothetical protein | - |
| PXH70_RS23165 (PXH70_23165) | 76302..76526 | - | 225 | WP_008324141 | TraR/DksA C4-type zinc finger protein | - |
| PXH70_RS23170 (PXH70_23170) | 76646..77185 | - | 540 | WP_008324154 | type IV conjugative transfer system lipoprotein TraV | traV |
| PXH70_RS23175 (PXH70_23175) | 77207..78601 | - | 1395 | WP_008324156 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PXH70_RS23180 (PXH70_23180) | 78588..79331 | - | 744 | WP_008324157 | type-F conjugative transfer system secretin TraK | traK |
| PXH70_RS23185 (PXH70_23185) | 79318..79884 | - | 567 | WP_008324158 | type IV conjugative transfer system protein TraE | traE |
| PXH70_RS23190 (PXH70_23190) | 79899..80204 | - | 306 | WP_008324160 | type IV conjugative transfer system protein TraL | traL |
| PXH70_RS23195 (PXH70_23195) | 80355..80705 | - | 351 | WP_008324162 | type IV conjugative transfer system pilin TraA | - |
| PXH70_RS23200 (PXH70_23200) | 80763..80987 | - | 225 | WP_008324165 | TraY domain-containing protein | - |
| PXH70_RS23205 (PXH70_23205) | 81074..81763 | - | 690 | WP_008324166 | hypothetical protein | - |
| PXH70_RS23210 (PXH70_23210) | 81952..82344 | - | 393 | WP_008324167 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PXH70_RS23215 (PXH70_23215) | 82776..83252 | + | 477 | WP_008324168 | transglycosylase SLT domain-containing protein | virB1 |
| PXH70_RS23220 (PXH70_23220) | 83299..83829 | - | 531 | WP_008324170 | antirestriction protein | - |
| PXH70_RS23225 (PXH70_23225) | 84456..85286 | - | 831 | Protein_94 | N-6 DNA methylase | - |
| PXH70_RS23230 (PXH70_23230) | 85329..85592 | - | 264 | WP_008324174 | hypothetical protein | - |
| PXH70_RS23235 (PXH70_23235) | 85678..86022 | - | 345 | WP_008324177 | hypothetical protein | - |
| PXH70_RS23240 (PXH70_23240) | 86069..86356 | - | 288 | WP_008324178 | hypothetical protein | - |
| PXH70_RS23245 (PXH70_23245) | 86410..86784 | - | 375 | WP_008324180 | hypothetical protein | - |
| PXH70_RS23250 (PXH70_23250) | 87411..87770 | - | 360 | WP_008324181 | hypothetical protein | - |
| PXH70_RS23255 (PXH70_23255) | 87832..88155 | - | 324 | WP_008324183 | hypothetical protein | - |
Host bacterium
| ID | 4787 | GenBank | NZ_CP118561 |
| Plasmid name | 2020CK-00216|unnamed1 | Incompatibility group | IncFIB |
| Plasmid size | 111328 bp | Coordinate of oriT [Strand] | 82602..82704 [-] |
| Host baterium | Enterobacter kobei strain 2020CK-00216 |
Cargo genes
| Drug resistance gene | aph(3')-Ia, qnrS1 |
| Virulence gene | - |
| Metal resistance gene | merE, merD, merA, merC, merP, merT, merR |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |