Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104331 |
| Name | oriT_KI4_B1|p1unnamed |
| Organism | Fructilactobacillus cliffordii strain KI4_B1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP097120 (32960..32999 [-], 40 nt) |
| oriT length | 40 nt |
| IRs (inverted repeats) | 18..23, 28..33 (AGGGAA..TTCCCT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 40 nt
>oriT_KI4_B1|p1unnamed
CAACTTCACGAGGAACGAGGGAACTGTTTCCCTTATGCTC
CAACTTCACGAGGAACGAGGGAACTGTTTCCCTTATGCTC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file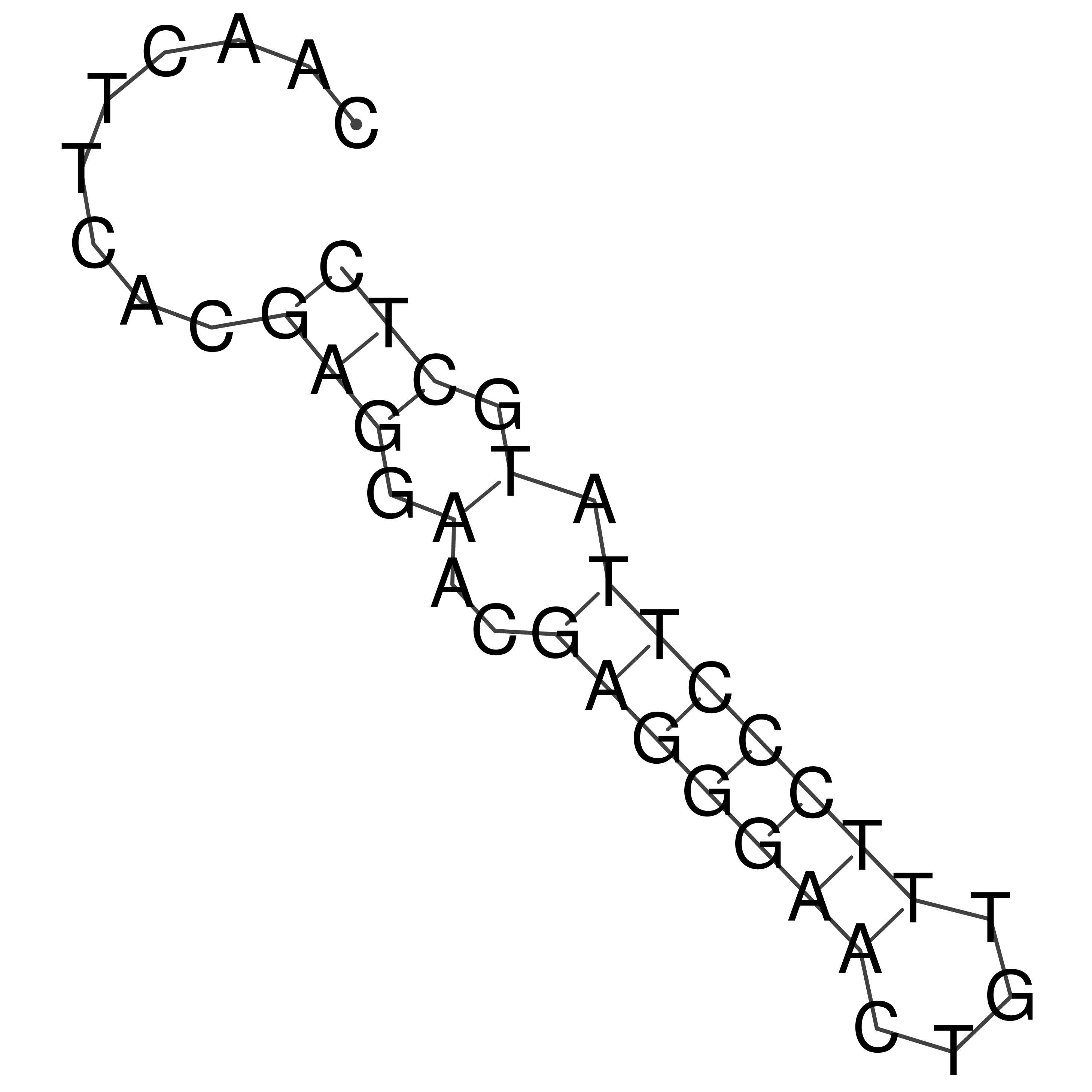
Relaxase
| ID | 3171 | GenBank | WP_252767520 |
| Name | mobP2_M3M40_RS07240_KI4_B1|p1unnamed |
UniProt ID | _ |
| Length | 664 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 664 a.a. Molecular weight: 77649.04 Da Isoelectric Point: 9.8833
>WP_252767520.1 MobP2 family relaxase [Fructilactobacillus cliffordii]
MKIKPIEKNAANVIFTTQFKTANQMTGAGKSFTKFVDYTDRDEAVDLGLKKDPKYKDFGGFIEYMDRDAA
TNIDVAKTRKLTSNFNEFKDHLNQDEVKELKDDLTKAQANNSNIYSNVVSFSTKFLKENKIYDDDKNELN
QEALRDIIRESMTDYISSTPLKNPIWWGDIHVNTEHVHVHVGMAEINPTTPEVVVGDHKEHKGKLPEHCF
RVFKKQVINKTLEFGKNKELDHKYSLEKDIGIQKQVLIDRVSDYDDLNNLMLALPENRRLWRYSSNAEDM
KPAKKILDRMVNQFKDGNPAYKMWLLKTEQLLTIESKNYGQSDSLNKRKQDMDKRLANRILKECRQVQDK
ELEISVSNIAQDFSDNSIQDNQNIIKVIRNRLKDRDNIEISGSKINDLKHEISIRKIALKKQIASKKIEE
IKEKDSKLNQISDYGLKPKQKAVINKLTTFIHQENLEKIELHELRQQKPWNLNDNEKRKLDKLSNKYVNA
ENVQIDTVNKDMVNNSRKRLQDINKLSDLNIIETKSFQILADEYPDIRIRSASRDLSILELKLKIKNNSS
NGRDNKENYERLKALSTSTNNANHKYKHKDLLVKPSYNKKNYKTFKKPKLFNQKLSHSLNNLTNDLNKNL
DSSIQNLSKQDNKEKRLQKDMLNDQRRDDDEYSR
MKIKPIEKNAANVIFTTQFKTANQMTGAGKSFTKFVDYTDRDEAVDLGLKKDPKYKDFGGFIEYMDRDAA
TNIDVAKTRKLTSNFNEFKDHLNQDEVKELKDDLTKAQANNSNIYSNVVSFSTKFLKENKIYDDDKNELN
QEALRDIIRESMTDYISSTPLKNPIWWGDIHVNTEHVHVHVGMAEINPTTPEVVVGDHKEHKGKLPEHCF
RVFKKQVINKTLEFGKNKELDHKYSLEKDIGIQKQVLIDRVSDYDDLNNLMLALPENRRLWRYSSNAEDM
KPAKKILDRMVNQFKDGNPAYKMWLLKTEQLLTIESKNYGQSDSLNKRKQDMDKRLANRILKECRQVQDK
ELEISVSNIAQDFSDNSIQDNQNIIKVIRNRLKDRDNIEISGSKINDLKHEISIRKIALKKQIASKKIEE
IKEKDSKLNQISDYGLKPKQKAVINKLTTFIHQENLEKIELHELRQQKPWNLNDNEKRKLDKLSNKYVNA
ENVQIDTVNKDMVNNSRKRLQDINKLSDLNIIETKSFQILADEYPDIRIRSASRDLSILELKLKIKNNSS
NGRDNKENYERLKALSTSTNNANHKYKHKDLLVKPSYNKKNYKTFKKPKLFNQKLSHSLNNLTNDLNKNL
DSSIQNLSKQDNKEKRLQKDMLNDQRRDDDEYSR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3082 | GenBank | WP_252767545 |
| Name | t4cp2_M3M40_RS07180_KI4_B1|p1unnamed |
UniProt ID | _ |
| Length | 966 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 966 a.a. Molecular weight: 110724.21 Da Isoelectric Point: 9.4979
>WP_252767545.1 VirD4-like conjugal transfer protein, CD1115 family [Fructilactobacillus cliffordii]
MFGKSLGKGKNRRTQAQGPWWSQSKKLLIRLLLITILVAITLVGYMTQVNYELFKYLRNLKLTSLLDPES
KVNHNPVTLYDIFNLKGFWIFNDLGYSGSIMIFWIVFVSIGLSAVWYRIFRSYITYNSYEYGNESFENNS
LVVKQYTNIPDRNKSFPGYGGIPVLHKNKFWGKLMQIYTYILAPKIKQSNENLDKVPGIPGINIQKRVQY
SFQKIKFLGKYMIDQATNNTLIYGITRSGKGETFVFPLIDILSRAMKKVSIFVNDPKGELARSSYFTLIR
RGYKVVIIDLSNMYKSASYNPLQVIIDYARKGYTDQVQMEVNKISTSIYQTKQSGNSDPFWANSSSNLLN
ALILAQIDLARRNNDWSKVTFFNVYQMLTQMAGKEVVVDSDERITDDVQNGTKKSMLTHYFNLMAQAPHD
PYRQMAMDAFSQSSFAGSETQGSIFSSMMEGIKIYQQQDVAKLTSKNTVDFKESAFPRRFSLKLMDTLQL
KTANVTFRDASGHLIESRKQDIDNLGYLNYALKAKLPRNFEITINVENHVYVFTGSIVRKQKVFGNPFKA
DVKVARQNIRILKNKKLKFIETKSTDNNYSDVQLKYSEQPVAVFFVTPPNNDAYNQIVSFAIDQSFNQMY
EMATQNGGKCFRRVHYVLDEFGNLPTVNSMNTKISIGLGQEILFDIVVQNDEQLSDKYDDKVANTIKANC
SNTLYILSNDDKTTELISKKLGKTTVMTRNHSITPDLNIGRDTLNTNQTAQDIMSSVELQHLMVGQQVLL
RANTRQSIYGNKIRNNPIFANGLKYEMPSRWQFLQDTFDTSTIMSDIPLKCIHSKMSLKDNAYNYFNNGM
NVSDNTELDKMNNLAGLSNSSTKISSSEKEIIKQVINEDDIRNAQEIENTYVDPLFTDEEISDESFLNKQ
LPKILNISKKQSSEGEEINEFLNSKTAIERINFIKNHNDQDFWNIFGITRENLDLD
MFGKSLGKGKNRRTQAQGPWWSQSKKLLIRLLLITILVAITLVGYMTQVNYELFKYLRNLKLTSLLDPES
KVNHNPVTLYDIFNLKGFWIFNDLGYSGSIMIFWIVFVSIGLSAVWYRIFRSYITYNSYEYGNESFENNS
LVVKQYTNIPDRNKSFPGYGGIPVLHKNKFWGKLMQIYTYILAPKIKQSNENLDKVPGIPGINIQKRVQY
SFQKIKFLGKYMIDQATNNTLIYGITRSGKGETFVFPLIDILSRAMKKVSIFVNDPKGELARSSYFTLIR
RGYKVVIIDLSNMYKSASYNPLQVIIDYARKGYTDQVQMEVNKISTSIYQTKQSGNSDPFWANSSSNLLN
ALILAQIDLARRNNDWSKVTFFNVYQMLTQMAGKEVVVDSDERITDDVQNGTKKSMLTHYFNLMAQAPHD
PYRQMAMDAFSQSSFAGSETQGSIFSSMMEGIKIYQQQDVAKLTSKNTVDFKESAFPRRFSLKLMDTLQL
KTANVTFRDASGHLIESRKQDIDNLGYLNYALKAKLPRNFEITINVENHVYVFTGSIVRKQKVFGNPFKA
DVKVARQNIRILKNKKLKFIETKSTDNNYSDVQLKYSEQPVAVFFVTPPNNDAYNQIVSFAIDQSFNQMY
EMATQNGGKCFRRVHYVLDEFGNLPTVNSMNTKISIGLGQEILFDIVVQNDEQLSDKYDDKVANTIKANC
SNTLYILSNDDKTTELISKKLGKTTVMTRNHSITPDLNIGRDTLNTNQTAQDIMSSVELQHLMVGQQVLL
RANTRQSIYGNKIRNNPIFANGLKYEMPSRWQFLQDTFDTSTIMSDIPLKCIHSKMSLKDNAYNYFNNGM
NVSDNTELDKMNNLAGLSNSSTKISSSEKEIIKQVINEDDIRNAQEIENTYVDPLFTDEEISDESFLNKQ
LPKILNISKKQSSEGEEINEFLNSKTAIERINFIKNHNDQDFWNIFGITRENLDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 4770 | GenBank | NZ_CP097120 |
| Plasmid name | KI4_B1|p1unnamed | Incompatibility group | - |
| Plasmid size | 41034 bp | Coordinate of oriT [Strand] | 32960..32999 [-] |
| Host baterium | Fructilactobacillus cliffordii strain KI4_B1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |