Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104237 |
| Name | oriT_pSymA |
| Organism | Sinorhizobium meliloti 1021 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_003037 (515639..515667 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file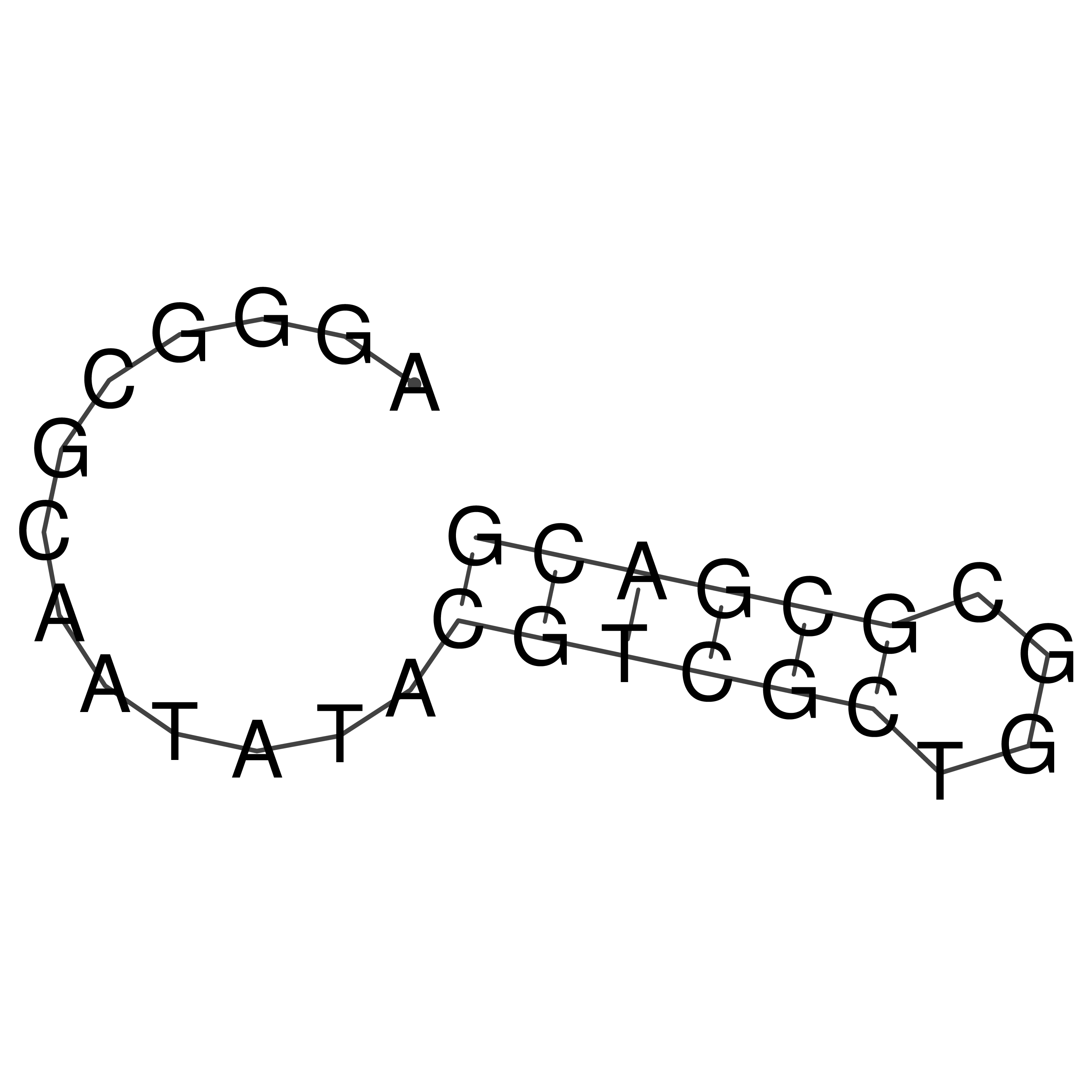
Relaxase
| ID | 3111 | GenBank | WP_010967486 |
| Name | traA_SM_RS27270_pSymA |
UniProt ID | Q92ZI0 |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169953.57 Da Isoelectric Point: 9.4213
>WP_010967486.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWENGREQLNRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTIVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALAALNALASDAAIEPRKLAEDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDQRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWENGREQLNRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTIVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARQERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALAALNALASDAAIEPRKLAEDLGKAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRGITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDQRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARSVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3015 | GenBank | WP_010967483 |
| Name | traG_SM_RS27255_pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70304.29 Da Isoelectric Point: 9.5501
>WP_010967483.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQERSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAP
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPEYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQERSASTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 711238..720817
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| SM_RS28200 (SMa1296) | 706855..707877 | - | 1023 | WP_010967682 | alcohol dehydrogenase AdhP | - |
| SM_RS28205 (SMa1297) | 708125..708802 | - | 678 | WP_010967683 | YoaK family protein | - |
| SM_RS28210 (SMa1299) | 709054..709665 | + | 612 | WP_010967684 | TetR/AcrR family transcriptional regulator | - |
| SM_RS28215 (SMa1301) | 709700..710926 | + | 1227 | WP_010967685 | MFS transporter | - |
| SM_RS28220 (SMa1302) | 711238..712227 | - | 990 | WP_010967686 | P-type DNA transfer ATPase VirB11 | virB11 |
| SM_RS28225 (SMa1303) | 712238..713410 | - | 1173 | WP_010967687 | type IV secretion system protein VirB10 | virB10 |
| SM_RS28230 (SMa1306) | 713420..714274 | - | 855 | WP_010967688 | P-type conjugative transfer protein VirB9 | virB9 |
| SM_RS28235 (SMa1308) | 714274..714945 | - | 672 | WP_010967689 | virB8 family protein | virB8 |
| SM_RS28240 (SMa1310) | 714950..715264 | - | 315 | WP_010967690 | hypothetical protein | - |
| SM_RS28245 (SMa1311) | 715261..716196 | - | 936 | WP_013845459 | type IV secretion system protein | virB6 |
| SM_RS28250 | 716202..716435 | - | 234 | WP_013845460 | EexN family lipoprotein | - |
| SM_RS28255 (SMa1313) | 716432..717133 | - | 702 | WP_010967692 | P-type DNA transfer protein VirB5 | virB5 |
| SM_RS28260 (SMa1315) | 717133..719529 | - | 2397 | WP_014528660 | VirB4 family type IV secretion system protein | virb4 |
| SM_RS28265 (SMa1318) | 719504..719845 | - | 342 | WP_010967694 | type IV secretion system protein VirB3 | virB3 |
| SM_RS28270 (SMa1319) | 719850..720149 | - | 300 | WP_010967695 | TrbC/VirB2 family protein | virB2 |
| SM_RS28275 (SMa1321) | 720146..720817 | - | 672 | WP_010967696 | lytic transglycosylase domain-containing protein | virB1 |
| SM_RS28280 (SMa1322) | 720821..721339 | - | 519 | WP_010967697 | hypothetical protein | - |
| SM_RS28285 (SMa1323) | 721436..721804 | + | 369 | WP_010967698 | transcriptional regulator | - |
| SM_RS28290 (SMa1325) | 722065..722316 | - | 252 | WP_010967699 | hypothetical protein | - |
| SM_RS28295 (SMa1326) | 722390..723037 | - | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| SM_RS28300 (SMa1327) | 723087..723935 | - | 849 | WP_010967701 | alpha/beta fold hydrolase | - |
Host bacterium
| ID | 4676 | GenBank | NC_003037 |
| Plasmid name | pSymA | Incompatibility group | - |
| Plasmid size | 1354226 bp | Coordinate of oriT [Strand] | 515639..515667 [+] |
| Host baterium | Sinorhizobium meliloti 1021 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP, nia, ncrA |
| Degradation gene | - |
| Symbiosis gene | nodD, fixN, fixO, fixU, nifB, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE, nifX, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nodM, fixS, fixG |
| Anti-CRISPR | AcrIIA9 |