Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104236 |
| Name | oriT_pSymB |
| Organism | Sinorhizobium meliloti 1021 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_003078 (725925..725953 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSymB
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file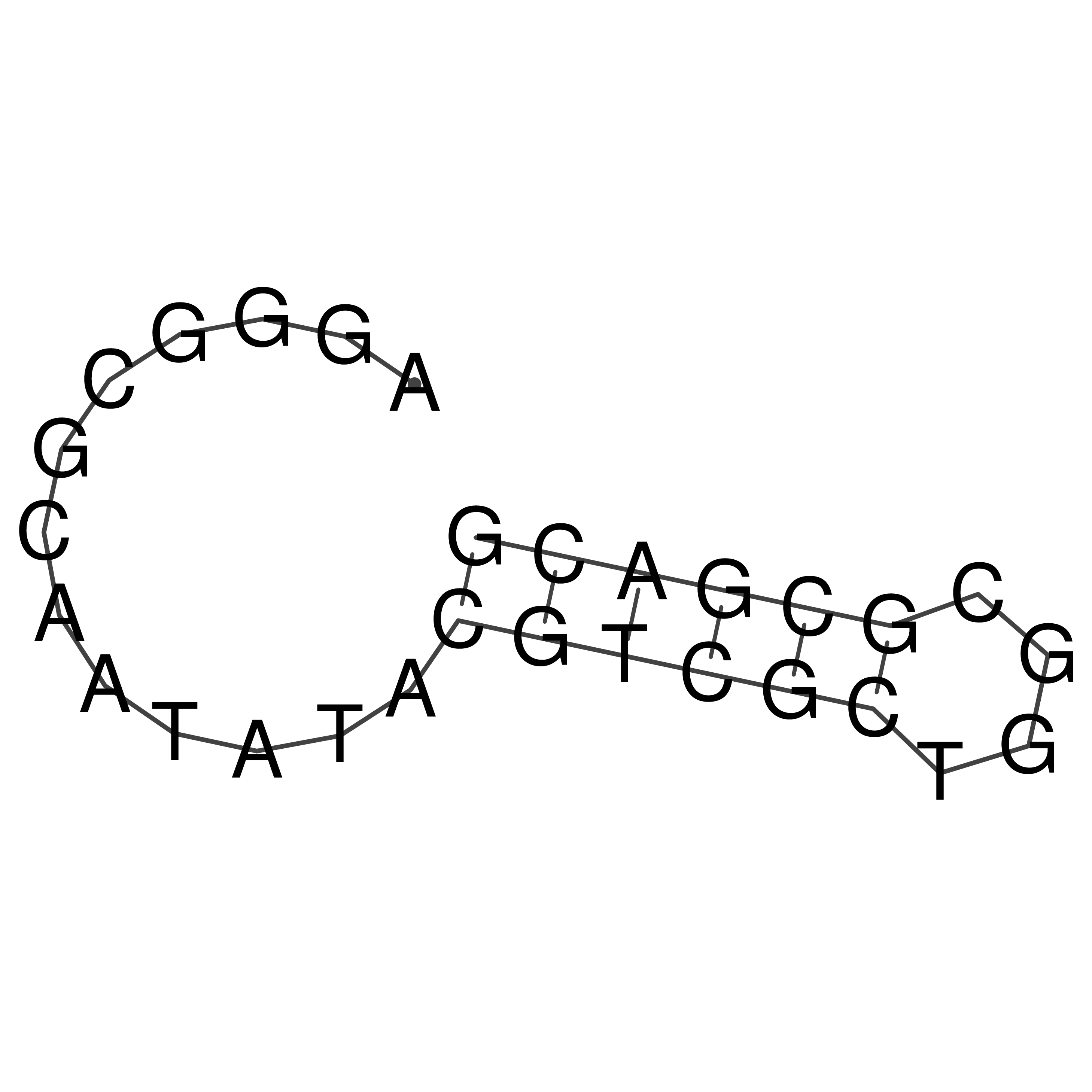
Relaxase
| ID | 3110 | GenBank | WP_010975542 |
| Name | traA_SM_RS20440_pSymB |
UniProt ID | Q92VN3 |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169833.61 Da Isoelectric Point: 9.6097
>WP_010975542.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIAG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALTGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEPKPEWGRKPRVDHAAGVTGELVEEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGAAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEGEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSEKIVSERRQRAINQSRGIRM
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIAG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDELVLLKPQQIDAETGKAKQPAVFTTREMLRLE
YAMAQSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDLEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLQRGDVLVIDEAGMVS
SQQMARVLKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAISERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALTGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEPKPEWGRKPRVDHAAGVTGELVEEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGAAEGDTITLRKDGVERVKVQVPIVDEQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEGEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSEKIVSERRQRAINQSRGIRM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3014 | GenBank | WP_167313204 |
| Name | tcpA_SM_RS24400_pSymB |
UniProt ID | _ |
| Length | 629 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 629 a.a. Molecular weight: 68084.03 Da Isoelectric Point: 4.7310
>WP_167313204.1 DNA translocase FtsK [Sinorhizobium meliloti]
MPVEIRPQPPTAISSLFRVVECRRPEPATAEPVTAEPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAI
VPASEPAPEAPVTRAAITMPAVIQRSSPSLPPIGAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLE
QNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNV
IGIELPNATRETVYFRELIESGDFQKTGCKLALCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINT
MILSLLYRLKPEECRLIMVDPKMLELSVYDGIPHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVR
NIDGYNQRAAAAREKGAPILATVQTGFEKGTGEPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGA
IQRLAQMARAAGIHLIMATQRPSVDVITGTIKANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHM
AGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPEYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYD
QAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVERMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
MPVEIRPQPPTAISSLFRVVECRRPEPATAEPVTAEPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAI
VPASEPAPEAPVTRAAITMPAVIQRSSPSLPPIGAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLE
QNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEFEPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNV
IGIELPNATRETVYFRELIESGDFQKTGCKLALCLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINT
MILSLLYRLKPEECRLIMVDPKMLELSVYDGIPHLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVR
NIDGYNQRAAAAREKGAPILATVQTGFEKGTGEPLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGA
IQRLAQMARAAGIHLIMATQRPSVDVITGTIKANFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHM
AGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPEYLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYD
QAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVERMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 4675 | GenBank | NC_003078 |
| Plasmid name | pSymB | Incompatibility group | - |
| Plasmid size | 1683333 bp | Coordinate of oriT [Strand] | 725925..725953 [+] |
| Host baterium | Sinorhizobium meliloti 1021 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7, AcrVIB |