Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104234 |
| Name | oriT_pF5.1a |
| Organism | Pararhizobium polonicum strain F5.1 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CM004502 (60064..60083 [-], 20 nt) |
| oriT length | 20 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 20 nt
>oriT_pF5.1a
ACGTATATTGCGCCCTCAAA
ACGTATATTGCGCCCTCAAA
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file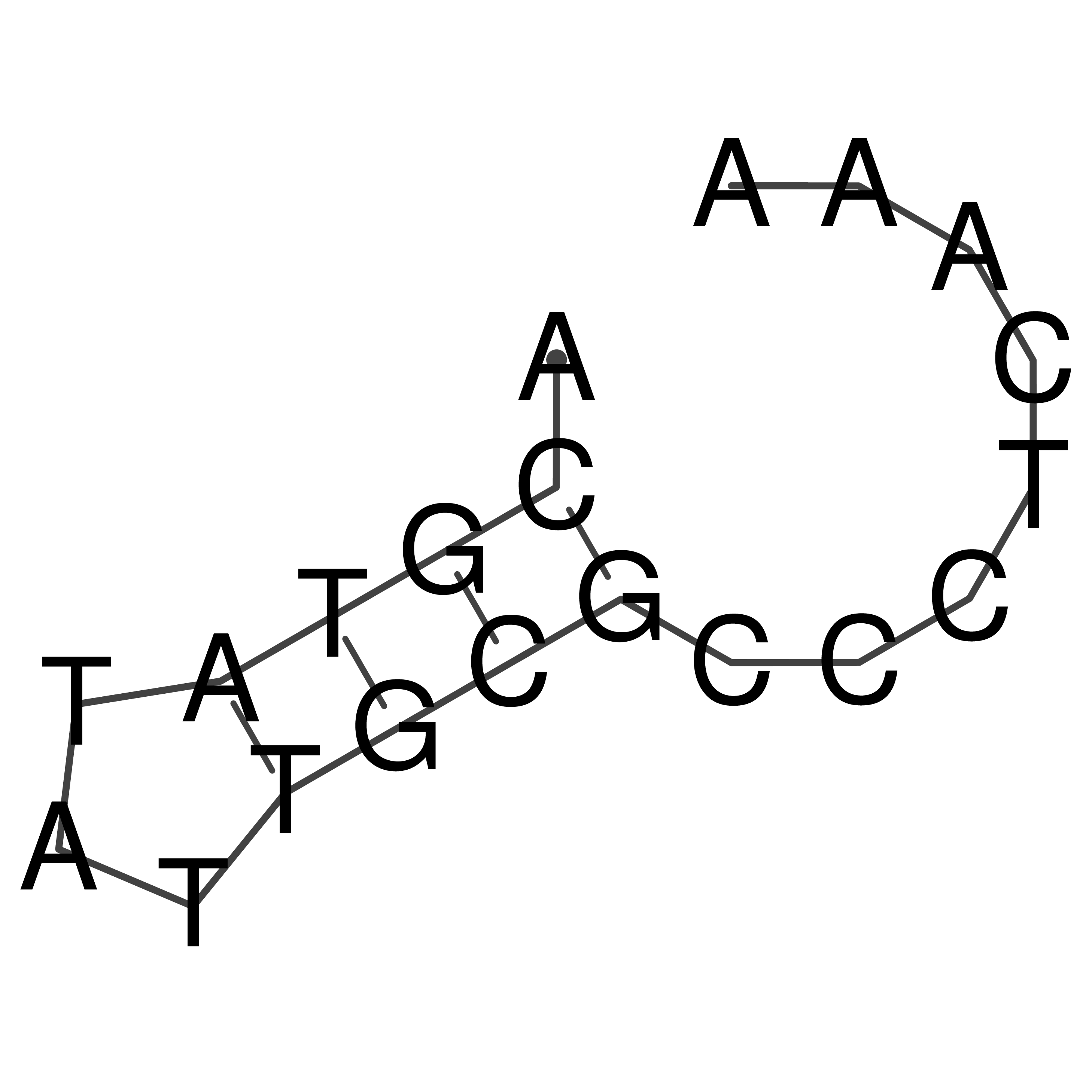
Relaxase
| ID | 3108 | GenBank | WP_068950593 |
| Name | traA_ADU59_RS00275_pF5.1a |
UniProt ID | A0A1C7P8U0 |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170730.99 Da Isoelectric Point: 7.9408
>WP_068950593.1 Ti-type conjugative transfer relaxase TraA [Pararhizobium polonicum]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDDQAGTSFSYRGGSRELVHEELAVPDEIPAWLKAAIDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVADNFTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGRKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKSWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLADPGLLLRQ
LGNERSTFDERDIAKALHRTVDDPVDFTNIRARLMASDDLVLLKPQELDSANGKVSEPAVFTTREILRIE
YDMAQSAEVLSKRTGFGIADAKVAVAVRQVETADPGKPFRLDAEQVDAVRHVTGAGAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWENGRDLLERGNVLVIDEAGMVS
SKQMARVLKVVEQAGAKTVLVGDAMQLQPIQAGAAFRAISERTGFAELAGVRRQREEWAREASRLFARGR
VEDGLDAYAQQGRVTEADTRAEIVERIVIDWTNARRDLLQKPGDEENVPRLRGDELLVLAHTNDDVKRLN
SSLRNVMMQEGALTDGREFQTARGLREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGSVVSTGDRR
GGPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRERVDL
YAAKEDFQPKPEWGRAARADHAAGVTGELVETGEAKFRPQDGDADESPYADVKTDDGTVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTVTVPVIDEKTGEKSFEERTVDRNVWTAKQVESAEARNERIEQEGHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYQAHARDFARRRGIDTLAGVAARQEEGAARRLAWVAEKREQVA
KLWERASVALGFAIERERRVSYSETRREPPTAEMSGGGKYLVPPTTVFARSVDEDARRAQLTSERWKERE
AILRPMLETIYRDPDQALIRLNTLSSETAVEPRQLTDALAAAPDRLGRLRGSDLVVDGRAGRGERHAAIA
ALSELLPLARAHATEFRRQAERFGIREEQRRAHMSLSIPVLSKPAMARLVEIEAVRDRGGDDAYKTAFTY
AAADRLLVQEVKAVNDALTARFGWSAFTAKADAVAERNTSERLPDDLTPERRQKLVRLFEVIRRFAEEQH
LAERQDRTKIVAGASVEVGKETVPVLPMLAAVTEFTTPVDAVARKRVLAIPHYRHHRSALAETATRIWRD
PAGAVARIEDLVVKGFAGERIAAAVGNDPAAYGALRGSDRMMDKLLAVGRERKEALQAVPEAASRVRSIS
ASYVSALDAETRTINEERRRMAVAVPGLSQAAEDALRQLVADVKTQKTQKEKGAKVDAAARSLDPDIGRE
FAAVSRALDERFGRNAILRGEADVINRVSPAQRRAFEAMRERLQVLQQTVRTHGSQEIISERQRRAIDRA
RGLTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDDQAGTSFSYRGGSRELVHEELAVPDEIPAWLKAAIDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENIALVREFVADNFTSKGMVADWVFHDKDG
NPHIHLMTTLRPLTEEGFGRKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETMKSWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLADPGLLLRQ
LGNERSTFDERDIAKALHRTVDDPVDFTNIRARLMASDDLVLLKPQELDSANGKVSEPAVFTTREILRIE
YDMAQSAEVLSKRTGFGIADAKVAVAVRQVETADPGKPFRLDAEQVDAVRHVTGAGAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWENGRDLLERGNVLVIDEAGMVS
SKQMARVLKVVEQAGAKTVLVGDAMQLQPIQAGAAFRAISERTGFAELAGVRRQREEWAREASRLFARGR
VEDGLDAYAQQGRVTEADTRAEIVERIVIDWTNARRDLLQKPGDEENVPRLRGDELLVLAHTNDDVKRLN
SSLRNVMMQEGALTDGREFQTARGLREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGSVVSTGDRR
GGPLLSVRLDNGRDVVISEDSYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRERVDL
YAAKEDFQPKPEWGRAARADHAAGVTGELVETGEAKFRPQDGDADESPYADVKTDDGTVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTVTVPVIDEKTGEKSFEERTVDRNVWTAKQVESAEARNERIEQEGHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYQAHARDFARRRGIDTLAGVAARQEEGAARRLAWVAEKREQVA
KLWERASVALGFAIERERRVSYSETRREPPTAEMSGGGKYLVPPTTVFARSVDEDARRAQLTSERWKERE
AILRPMLETIYRDPDQALIRLNTLSSETAVEPRQLTDALAAAPDRLGRLRGSDLVVDGRAGRGERHAAIA
ALSELLPLARAHATEFRRQAERFGIREEQRRAHMSLSIPVLSKPAMARLVEIEAVRDRGGDDAYKTAFTY
AAADRLLVQEVKAVNDALTARFGWSAFTAKADAVAERNTSERLPDDLTPERRQKLVRLFEVIRRFAEEQH
LAERQDRTKIVAGASVEVGKETVPVLPMLAAVTEFTTPVDAVARKRVLAIPHYRHHRSALAETATRIWRD
PAGAVARIEDLVVKGFAGERIAAAVGNDPAAYGALRGSDRMMDKLLAVGRERKEALQAVPEAASRVRSIS
ASYVSALDAETRTINEERRRMAVAVPGLSQAAEDALRQLVADVKTQKTQKEKGAKVDAAARSLDPDIGRE
FAAVSRALDERFGRNAILRGEADVINRVSPAQRRAFEAMRERLQVLQQTVRTHGSQEIISERQRRAIDRA
RGLTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 3011 | GenBank | WP_068950587 |
| Name | traG_ADU59_RS00260_pF5.1a |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70352.05 Da Isoelectric Point: 9.5787
>WP_068950587.1 Ti-type conjugative transfer system protein TraG [Pararhizobium polonicum]
MTIRAKPHPSLLLTFVPITVTAFAAYVVGWRWPELAAGMTGKTQYWFLRASPVPVLLFGPLAGLLTVWSL
PLHRRRPIAMATLICFLSVAGFYALREFGRLSPSVQSGGVTWNQALSFIDMVAVVGAVVGFMATAVSARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDKEIVHELPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQHSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKPSDLTGGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFHRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRSGRAIYFRRKDMNAEAK
VNRFVKP
MTIRAKPHPSLLLTFVPITVTAFAAYVVGWRWPELAAGMTGKTQYWFLRASPVPVLLFGPLAGLLTVWSL
PLHRRRPIAMATLICFLSVAGFYALREFGRLSPSVQSGGVTWNQALSFIDMVAVVGAVVGFMATAVSARI
STVVSEPVKRAKRGTFGDADWLSMSAAGRLFPDDGEIVVGERYRVDKEIVHELPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSKQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPETSVLAMLRDVQQHSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALVC
GNTFKPSDLTGGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFHRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIVVQGHSPIRSGRAIYFRRKDMNAEAK
VNRFVKP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 170704..180287
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| ADU59_RS00760 (ADU59_00770) | 165791..167173 | - | 1383 | WP_158515689 | MFS transporter | - |
| ADU59_RS00765 (ADU59_00775) | 167254..168189 | - | 936 | WP_068950759 | polysaccharide deacetylase | - |
| ADU59_RS00770 (ADU59_00780) | 168191..169129 | - | 939 | WP_338012923 | polysaccharide deacetylase family protein | - |
| ADU59_RS00775 (ADU59_00785) | 169254..169949 | + | 696 | WP_068950761 | TetR/AcrR family transcriptional regulator | - |
| ADU59_RS29905 | 169953..170073 | - | 121 | Protein_161 | glutamine ABC transporter ATP-binding protein GlnQ | - |
| ADU59_RS29430 | 170342..170485 | - | 144 | Protein_162 | cold-shock protein | - |
| ADU59_RS00780 (ADU59_00790) | 170704..171705 | - | 1002 | WP_068950763 | P-type DNA transfer ATPase VirB11 | virB11 |
| ADU59_RS00785 (ADU59_00795) | 171714..172877 | - | 1164 | WP_068950765 | type IV secretion system protein VirB10 | virB10 |
| ADU59_RS00790 (ADU59_00800) | 172892..173746 | - | 855 | WP_162493764 | P-type conjugative transfer protein VirB9 | virB9 |
| ADU59_RS00795 (ADU59_00805) | 173743..174414 | - | 672 | WP_068950767 | virB8 family protein | virB8 |
| ADU59_RS00800 (ADU59_00810) | 174418..174684 | - | 267 | WP_068950769 | hypothetical protein | - |
| ADU59_RS00805 (ADU59_00815) | 174720..175655 | - | 936 | WP_068950771 | type IV secretion system protein | virB6 |
| ADU59_RS00810 (ADU59_00820) | 175659..175892 | - | 234 | WP_068950773 | EexN family lipoprotein | - |
| ADU59_RS00815 (ADU59_00825) | 175889..176587 | - | 699 | WP_068950775 | P-type DNA transfer protein VirB5 | virB5 |
| ADU59_RS00820 (ADU59_00830) | 176587..178968 | - | 2382 | WP_068950777 | VirB4 family type IV secretion system protein | virb4 |
| ADU59_RS00825 (ADU59_00835) | 178961..179302 | - | 342 | WP_068950779 | type IV secretion system protein VirB3 | virB3 |
| ADU59_RS00830 (ADU59_00840) | 179305..179604 | - | 300 | WP_068950781 | TrbC/VirB2 family protein | virB2 |
| ADU59_RS00835 (ADU59_00845) | 179601..180287 | - | 687 | WP_068950783 | type IV secretion system protein VirB1 | virB1 |
| ADU59_RS31070 (ADU59_00850) | 180290..180829 | - | 540 | WP_068950785 | hypothetical protein | - |
| ADU59_RS00845 (ADU59_00855) | 180923..181285 | + | 363 | WP_068950787 | MarR family transcriptional regulator | - |
Host bacterium
| ID | 4673 | GenBank | NZ_CM004502 |
| Plasmid name | pF5.1a | Incompatibility group | - |
| Plasmid size | 181327 bp | Coordinate of oriT [Strand] | 60064..60083 [-] |
| Host baterium | Pararhizobium polonicum strain F5.1 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | bphC, genH |
| Symbiosis gene | - |
| Anti-CRISPR | - |