Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104233 |
| Name | oriT_pTEF2 |
| Organism | Enterococcus faecalis V583 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NC_004671 (29892..29941 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 34..35 |
| Conserved sequence flanking the nic site |
GCTTGCAAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pTEF2
AATATCGCAACATGGTACCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
AATATCGCAACATGGTACCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file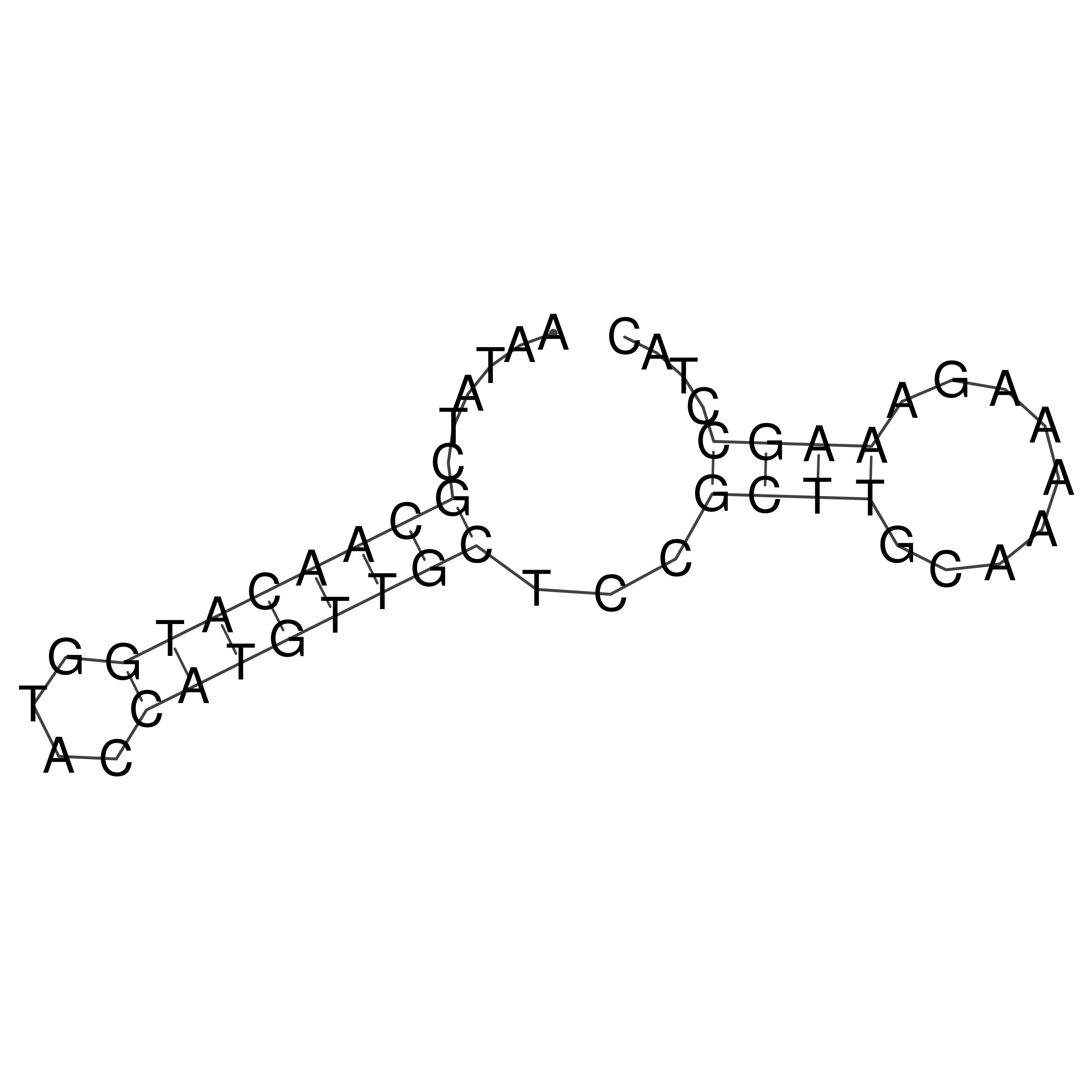
Relaxase
| ID | 3106 | GenBank | WP_010774278 |
| Name | Relaxase_EF_RS15915_pTEF2 |
UniProt ID | Q82YL3 |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65308.93 Da Isoelectric Point: 6.1201
>WP_010774278.1 relaxase/mobilization nuclease domain-containing protein [Enterococcus faecalis]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLANGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLANGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q82YL3 |
Auxiliary protein
| ID | 1353 | GenBank | WP_002367696 |
| Name | WP_002367696_pTEF2 |
UniProt ID | Q82YL4 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 14042.10 Da Isoelectric Point: 9.9724
>WP_002367696.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Bacteria]
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
MENKQKLKRPIQRKIRFSKEENELINRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKEQS
Protein domains
Predicted by InterproScan.
Protein structure
| Source | ID | Structure |
|---|---|---|
| AlphaFold DB | Q82YL4 |
T4CP
| ID | 3009 | GenBank | WP_002425085 |
| Name | t4cp2_EF_RS15895_pTEF2 |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70145.14 Da Isoelectric Point: 6.1811
>WP_002425085.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEGEKTTGEDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEGEKTTGEDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 9960..27248
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| EF_RS15805 (EF_B0007) | 5822..6202 | + | 381 | WP_002365885 | PrgR | - |
| EF_RS16875 | 6253..6474 | + | 222 | WP_016616022 | hypothetical protein | - |
| EF_RS15810 | 6776..7081 | + | 306 | Protein_6 | hypothetical protein | - |
| EF_RS15815 (EF_B0010) | 7092..9767 | + | 2676 | WP_011109619 | surface exclusion protein SEA1/PrgA | - |
| EF_RS15820 (EF_B0011) | 9960..13877 | + | 3918 | WP_002395058 | LPXTG-anchored aggregation substance PrgB/Asc10 | prgB |
| EF_RS15825 | 13939..14295 | + | 357 | WP_002367672 | pheromone response system RNA-binding regulator PrgU | - |
| EF_RS15830 (EF_B0012) | 14323..15180 | + | 858 | WP_002367673 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| EF_RS15835 (EF_B0013) | 15222..16121 | + | 900 | WP_011109620 | hypothetical protein | - |
| EF_RS15840 (EF_B0014) | 16141..16575 | + | 435 | WP_002365857 | hypothetical protein | - |
| EF_RS15845 (EF_B0015) | 16622..16849 | + | 228 | WP_002365858 | hypothetical protein | - |
| EF_RS15850 (EF_B0016) | 16863..17159 | + | 297 | WP_002387740 | hypothetical protein | - |
| EF_RS15855 (EF_B0017) | 17174..17974 | + | 801 | WP_010774271 | type IV secretion system protein | prgHb |
| EF_RS15860 (EF_B0018) | 17976..18329 | + | 354 | WP_010774272 | PrgI family protein | prgIc |
| EF_RS15865 (EF_B0019) | 18283..20673 | + | 2391 | WP_010774273 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| EF_RS15870 (EF_B0020) | 20685..23300 | + | 2616 | WP_010774274 | phage tail tip lysozyme | prgK |
| EF_RS15875 (EF_B0021) | 23324..23950 | + | 627 | WP_002395055 | hypothetical protein | prgL |
| EF_RS15880 (EF_B0022) | 23937..24182 | + | 246 | WP_002365866 | hypothetical protein | - |
| EF_RS15885 (EF_B0023) | 24166..24774 | + | 609 | WP_010774275 | hypothetical protein | - |
| EF_RS15890 (EF_B0024) | 24934..25419 | + | 486 | WP_010774276 | DUF3801 domain-containing protein | gbs1365 |
| EF_RS15895 (EF_B0025) | 25419..27248 | + | 1830 | WP_002425085 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| EF_RS15900 (EF_B0026) | 27298..29457 | + | 2160 | WP_010774277 | ArdC-like ssDNA-binding domain-containing protein | - |
| EF_RS15905 (EF_B0027) | 29490..29762 | + | 273 | WP_002362409 | LtrD | - |
| EF_RS15910 (EF_B0029) | 30008..30364 | + | 357 | WP_002367696 | plasmid mobilization relaxosome protein MobC | - |
| EF_RS15915 (EF_B0030) | 30365..32050 | + | 1686 | WP_010774278 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 4672 | GenBank | NC_004671 |
| Plasmid name | pTEF2 | Incompatibility group | - |
| Plasmid size | 57660 bp | Coordinate of oriT [Strand] | 29892..29941 [+] |
| Host baterium | Enterococcus faecalis V583 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA21, AcrIIA9 |