Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104215 |
| Name | oriT_pV386 |
| Organism | Enterococcus faecalis strain V386 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_MZ603802 (14803..14832 [+], 30 nt) |
| oriT length | 30 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 30 nt
>oriT_pV386
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
AACGTAAGTTAGTGGGTATAAGTGCGCCCT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file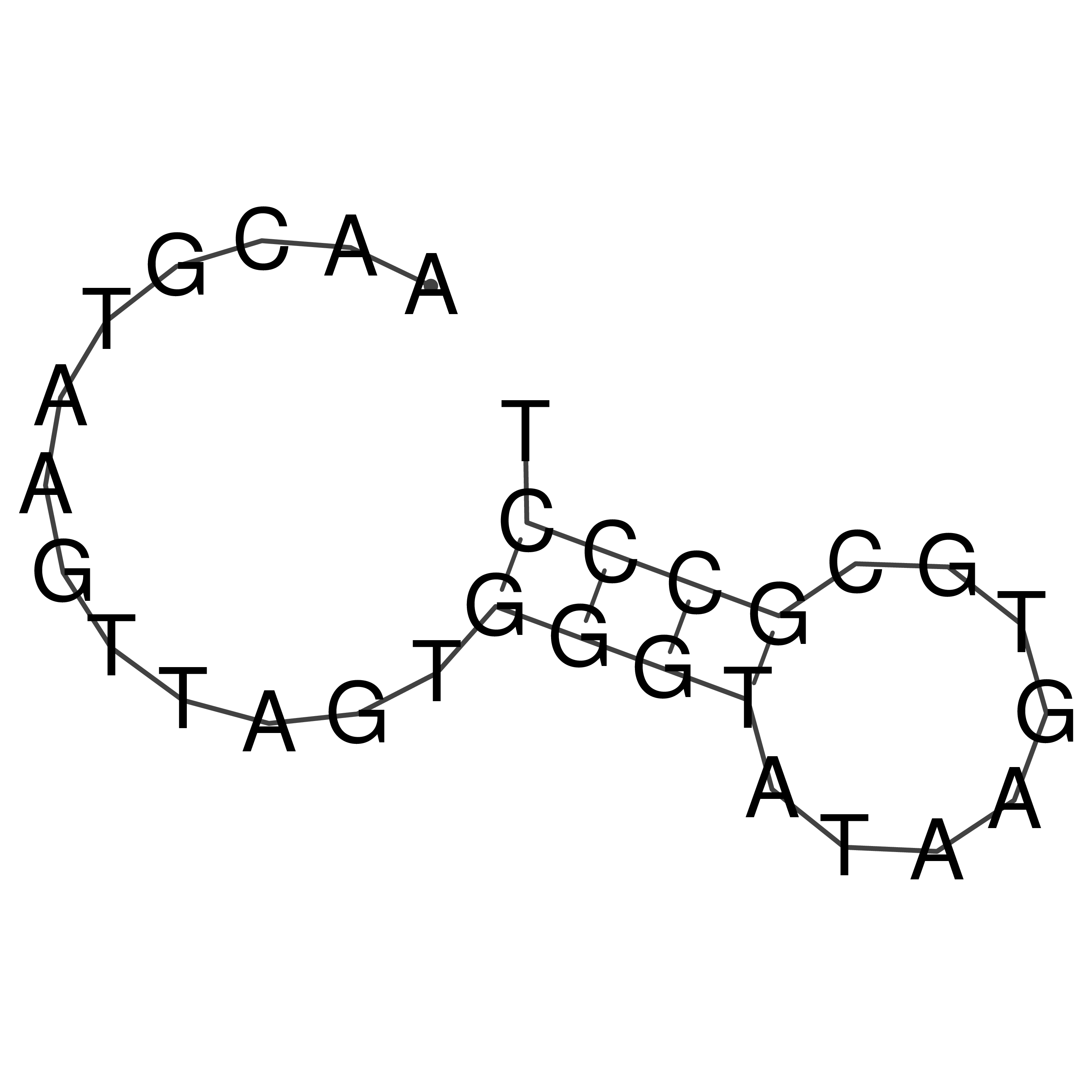
Relaxase
| ID | 3092 | GenBank | WP_224993994 |
| Name | mobQ_MGJ71_RS00075_pV386 |
UniProt ID | _ |
| Length | 664 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 664 a.a. Molecular weight: 77765.02 Da Isoelectric Point: 9.5003
>WP_224993994.1 MULTISPECIES: MobQ family relaxase [Enterococcus]
MIQLAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTIKPESFILKPDYVPDEFLDR
QTLWNKMELAEKSPNAQLCRELNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHI
MLTMREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKD
RNIDQQITEKSHAELGKKELPKIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQ
DQNFTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISS
ERDMLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVIEKEE
SKKIFKEKPFQTSRYLDSKIKQVEDSITKEKNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVS
IDSVIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMK
HIDELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKNEPEKNEVSVKMFQFAKSINRL
LSGNQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
MIQLAIFHLSMTIAKREGGKRSLIAMASYRSGEKLYSELYEKTNLYNHRTIKPESFILKPDYVPDEFLDR
QTLWNKMELAEKSPNAQLCRELNVALPIELNNSDQRMLIEDFVKDNFVSEGMIADVAIHRDDENNPHAHI
MLTMREVDSEGNILNKRKRIPKLDENGNQIFNEKGQRVTVSIKTNDWDRKSLVSEIRKDWADKVNQYLKD
RNIDQQITEKSHAELGKKELPKIHEGFYSKKLEDKGVISELKRKNLEIQSYNDILAELDKLESQEKVLKQ
DQNFTLKFEKTFSPLEKGELKNLSKELKLFINDENIDKRLGELKRWENSLIFNNKMEIQKQRLMLSKISS
ERDMLTKANEILDKQAERFFKKSYPSLNIDKFSNHEVRAMVNETIFRKQLLNKDQLAEVIYNERVIEKEE
SKKIFKEKPFQTSRYLDSKIKQVEDSITKEKNPERKEILSIKKEKLIGIKQGLIEYVQSEVERKFDKNVS
IDSVIEGEMLLAKADYYKTTDFSKVEGVARFSSEEINSMLEQSKGFLTNIQTVKIPNDCQGVFFVQDSMK
HIDELSPLAKHNLKKVVNRNAYLPDSDKIELSKEIENTNKDQSQELDKNEPEKNEVSVKMFQFAKSINRL
LSGNQLQKKRNLDKLIKQTKAKKNQSLQRNIPLR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2994 | GenBank | WP_224993997 |
| Name | t4cp2_MGJ71_RS00030_pV386 |
UniProt ID | _ |
| Length | 551 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 551 a.a. Molecular weight: 63110.11 Da Isoelectric Point: 4.7040
>WP_224993997.1 MULTISPECIES: VirD4-like conjugal transfer protein, CD1115 family [Enterococcus]
MTSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFRTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKETIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
MTSLLAESDGLILGKFQSGNVVIQPEDSKISNRNIFVVGGPGSFKTQSYVLPNVVNNRSTSIVVTDPKGE
IYELTNEIKKAQGFRTVVINFKDFLLSSRYNPLLYIRKSNDTNKIANVIVSAKNDPKRKDFWFNAQLNLL
NTLIKYVYFEYEPSARTIEGILDFLEEFDPRYNEEGVSELDEQFERLPDGHEAKRSYYLGFRQAQSEARP
NIVISLLTTLQDFVDKEVSEFTSTNDFFFEELGTSKICLYVLISPLDRTWDGLVNLFFQQMFTELYFLGD
KHNAKLPVPLVMLLDEFVNLGYFPTYENFLATCRGYRISVSTILQSLPQGFELYGDKKFKAIIGNHAIKI
CLGGVEETTAEYFSRQVNDTTIKVYTGGTSESKTSAKTTNRSGSKSESYGYQKRRLITEGEVINLQQEEN
GRKSIVLIDGKPYMLRKTPQFELFGNLLKKHEISQQDYISSQTEFAKETIEELEKQHKQRKVQLASAPIF
HEKKQEDDLNKKEINLPEQPIESEVTEEEEEKELLMSDILSSFDEGIEENTTNNDNSELPF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 49..12337
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| MGJ71_RS00005 | 49..861 | - | 813 | WP_118368352 | LPXTG cell wall anchor domain-containing protein | prgC |
| MGJ71_RS00010 | 921..1289 | - | 369 | WP_000516429 | hypothetical protein | - |
| MGJ71_RS00015 | 1318..2289 | - | 972 | WP_002425009 | hypothetical protein | - |
| MGJ71_RS00020 | 2306..3250 | - | 945 | WP_194187774 | conjugal transfer protein TrbL family protein | trsL |
| MGJ71_RS00025 | 3250..4173 | - | 924 | WP_224993998 | hypothetical protein | traP |
| MGJ71_RS00030 | 4191..5846 | - | 1656 | WP_224993997 | VirD4-like conjugal transfer protein, CD1115 family | - |
| MGJ71_RS00035 | 5839..6270 | - | 432 | WP_224993996 | hypothetical protein | - |
| MGJ71_RS00040 | 6275..6826 | - | 552 | WP_224993995 | hypothetical protein | - |
| MGJ71_RS00045 | 6839..7948 | - | 1110 | WP_002425005 | lysozyme family protein | orf14 |
| MGJ71_RS00050 | 7970..9322 | - | 1353 | WP_015543616 | hypothetical protein | trsF |
| MGJ71_RS00055 | 9336..11297 | - | 1962 | WP_010783404 | hypothetical protein | virb4 |
| MGJ71_RS00060 | 11308..11937 | - | 630 | WP_010783405 | hypothetical protein | trsD |
| MGJ71_RS00065 | 11954..12337 | - | 384 | WP_010783406 | hypothetical protein | trsC |
| MGJ71_RS00070 | 12356..12688 | - | 333 | WP_000713503 | CagC family type IV secretion system protein | - |
| MGJ71_RS00075 | 12712..14697 | - | 1986 | WP_194187770 | MobQ family relaxase | - |
| MGJ71_RS00080 | 14989..15288 | + | 300 | WP_002325624 | hypothetical protein | - |
| MGJ71_RS00085 | 15291..15548 | + | 258 | WP_000002668 | hypothetical protein | - |
| MGJ71_RS00090 | 15571..15876 | - | 306 | WP_070544069 | hypothetical protein | - |
| MGJ71_RS00095 | 15910..16407 | - | 498 | WP_010713926 | DnaJ domain-containing protein | - |
| MGJ71_RS00100 | 16427..16744 | - | 318 | WP_002338433 | hypothetical protein | - |
Host bacterium
| ID | 4654 | GenBank | NZ_MZ603802 |
| Plasmid name | pV386 | Incompatibility group | - |
| Plasmid size | 33480 bp | Coordinate of oriT [Strand] | 14803..14832 [+] |
| Host baterium | Enterococcus faecalis strain V386 |
Cargo genes
| Drug resistance gene | Cfr(D), poxtA, fexA |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |