Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104138 |
| Name | oriT_pJXZ076-99K |
| Organism | Enterococcus faecalis strain CQJXZ21-076 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP117504 (96487..96536 [+], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | _ |
| Location of nic site | 34..35 |
| Conserved sequence flanking the nic site |
GCTTGCAAAA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_pJXZ076-99K
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
AATATCGCAACATGCTAGCATGTTGCTCCGCTTGCAAAAAGAAAGCCTAC
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file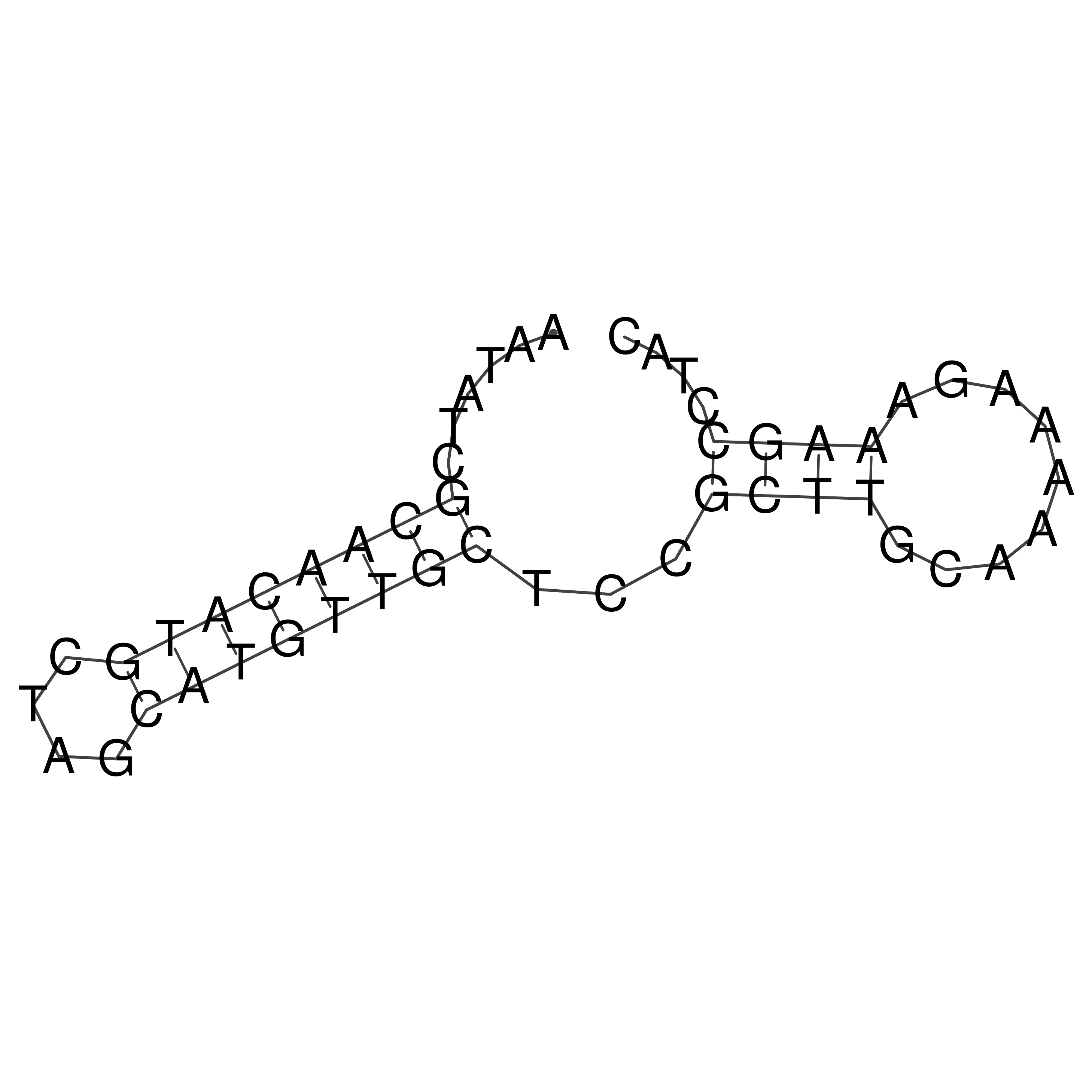
Relaxase
| ID | 3042 | GenBank | WP_010784826 |
| Name | Relaxase_PSC77_RS14140_pJXZ076-99K |
UniProt ID | A0A8E7CAC2 |
| Length | 561 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 561 a.a. Molecular weight: 65352.94 Da Isoelectric Point: 6.0135
>WP_010784826.1 MULTISPECIES: relaxase/mobilization nuclease domain-containing protein [Enterococcus]
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
MVYTKHFVIHTFDKLNNACSYIENAEKTEVTNDNPSEHLEHLFQYIVNDDKTYMKKLVSGHGIVDPTNPY
EEFKLTKLQAAIQRKIGYTFDPKSERLLPPTLTELEKGNAVLAHHLIQSFSPEDDLTPEKIHEIGYNTVM
ELTGGEYEFVIATHVDKEHLHNHIIFSSTNLKTGKAFRWQKGTKKVFEQISDKIAAKEGAKIIEKSPKQT
HTKYTKWQTESIYKQKIKSRLDFLLEQSSSINDFLEKAEALNLSADFSNKWTTYRLLDEPQIKNTRSRSL
SKSDPTRYNYEKIIERLKENKNVLTVEECVKRYEEKNEQEKNNFDYQFTIEPWQLSHKTERGYYINIDYG
YGNSGKLFVGGYKVDPLDNGNYNIYIKRNEYFYFMSDKDATKSKYLTGASLIKQLRLYNGQTPLKREPVM
QTIDELVSAINFLAANEIEDTRQLKLLEEKLEVAFLEAEKTLETLDEKMLELHQLSNLLLENELQGDPEL
IQKKLKTLLPDATLAEFSYEDVRGEIEAIKTSQTLLESKLERTRNEINQLHEIQAVQEKEPEKQTDIKPK
L
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1310 | GenBank | WP_002393737 |
| Name | WP_002393737_pJXZ076-99K |
UniProt ID | A0A8E7CBI6 |
| Length | 118 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 118 a.a. Molecular weight: 13965.01 Da Isoelectric Point: 9.6545
>WP_002393737.1 MULTISPECIES: plasmid mobilization relaxosome protein MobC [Enterococcus]
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKDQS
MENKQKLKRPIQRIVRLSEEENNLIKRKIEESFFPNFQNFALHLLIQGEIRHVDYSELNRLTTEIHKIGI
NINQMARLANQFHEISSEDIKDLTDKVQSLNALVQSELNKLIKRKDQS
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2930 | GenBank | WP_002401458 |
| Name | t4cp2_PSC77_RS14120_pJXZ076-99K |
UniProt ID | _ |
| Length | 609 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 609 a.a. Molecular weight: 70173.19 Da Isoelectric Point: 6.1807
>WP_002401458.1 VirD4-like conjugal transfer protein, CD1115 family [Enterococcus faecalis]
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
MQTYKKELKPYLYLGIFLFVIGFGIGNFFWLLPGTDYFSKTNYLFTKDILTYIEQTFYRFFITPSLLGFS
VGILGFLLGLLMYVRDNDRGIYRHGEEYGSARFATPAEMKKYEDPIPENNIIVSKHVKISLFNKRLPIKL
QKNKNIAILGDSGAAKTLAFIKTNLMQRHASFITTDPDGGILPEIGLLLKKGKYKIKVLDLNTLSNSDTF
NVFEYMHSELDVDRVLEAVTEATKEVEKTTGDDFWIKAEGLLVRSLIAYLWFDGRRNDYTPHLGMIADML
RHLKRKDKNVPSPVEEWFEELNEAIPDNYAYRQWTLFNNLYESETRASVLGIAAARYSVFDHEEVVNMIR
TDTMDIDSWNEEKTAVFIAIPETNTSFNFIAALMFATVGEVLRNKSDQVRLGKRVLEEGKELLHVRFLID
EFANIGRIPHFEKMLTTFRKREMSFTIVLQSLNQLQTLYRNGWQNILNGCACLLYLGGDEEETTKYLSAR
AGKQTLSIRKHSMNKGNQGGGSENRDKYGRDLIDRSEVALIGGDECLVFISKEHVFKDKKFYAFQHDRAD
ELATGPTDDKWYTWRRYMTDEEAILDKISATDIIDHGEITSELKEETHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 75100..93842
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PSC77_RS14005 (PSC77_14005) | 70215..70595 | + | 381 | WP_002367664 | hypothetical protein | - |
| PSC77_RS14010 (PSC77_14010) | 71166..71471 | + | 306 | WP_126263119 | hypothetical protein | - |
| PSC77_RS14015 (PSC77_14015) | 71482..74154 | + | 2673 | WP_273982070 | surface exclusion protein SEA1/PrgA | - |
| PSC77_RS14020 (PSC77_14020) | 74233..74427 | + | 195 | WP_002415094 | hypothetical protein | - |
| PSC77_RS14025 (PSC77_14025) | 74700..74888 | + | 189 | Protein_87 | hypothetical protein | - |
| PSC77_RS14030 (PSC77_14030) | 75100..79017 | + | 3918 | WP_104866339 | LPXTG-anchored aggregation substance PrgB/Asc10 | prgB |
| PSC77_RS14035 (PSC77_14035) | 79079..79435 | + | 357 | WP_002367672 | pheromone response system RNA-binding regulator PrgU | - |
| PSC77_RS14040 (PSC77_14040) | 79463..80320 | + | 858 | WP_002367673 | pheromone response LPXTG-anchored adhesin PrgC | prgC |
| PSC77_RS14045 (PSC77_14045) | 80362..81261 | + | 900 | WP_002409287 | hypothetical protein | - |
| PSC77_RS14050 (PSC77_14050) | 81281..81715 | + | 435 | WP_002365857 | hypothetical protein | - |
| PSC77_RS14055 (PSC77_14055) | 81762..81989 | + | 228 | WP_002365858 | hypothetical protein | - |
| PSC77_RS14060 (PSC77_14060) | 82003..82299 | + | 297 | WP_002387740 | hypothetical protein | - |
| PSC77_RS14065 (PSC77_14065) | 82314..83114 | + | 801 | WP_002393746 | hypothetical protein | prgHb |
| PSC77_RS14070 (PSC77_14070) | 83116..83469 | + | 354 | WP_002367919 | PrgI family mobile element protein | prgIc |
| PSC77_RS14075 (PSC77_14075) | 83423..85813 | + | 2391 | WP_002365863 | VirB4-like conjugal transfer ATPase, CD1110 family | virb4 |
| PSC77_RS14080 (PSC77_14080) | 85825..88440 | + | 2616 | WP_002393744 | phage tail tip lysozyme | prgK |
| PSC77_RS14085 (PSC77_14085) | 88464..89090 | + | 627 | WP_002386490 | hypothetical protein | prgL |
| PSC77_RS14090 (PSC77_14090) | 89077..89322 | + | 246 | WP_002365866 | hypothetical protein | - |
| PSC77_RS14095 (PSC77_14095) | 89306..89914 | + | 609 | WP_016618746 | hypothetical protein | - |
| PSC77_RS14100 (PSC77_14100) | 90028..90783 | + | 756 | WP_077932946 | Fic family protein | - |
| PSC77_RS14105 (PSC77_14105) | 90800..91258 | + | 459 | WP_077932945 | single-stranded DNA-binding protein | - |
| PSC77_RS14110 (PSC77_14110) | 91304..91486 | + | 183 | WP_002367689 | hypothetical protein | - |
| PSC77_RS14115 (PSC77_14115) | 91528..92013 | + | 486 | WP_002386493 | PcfB family protein | gbs1365 |
| PSC77_RS14120 (PSC77_14120) | 92013..93842 | + | 1830 | WP_002401458 | VirD4-like conjugal transfer protein, CD1115 family | virb4 |
| PSC77_RS14125 (PSC77_14125) | 93893..96052 | + | 2160 | WP_010784825 | ArdC-like ssDNA-binding domain-containing protein | - |
| PSC77_RS14130 (PSC77_14130) | 96085..96357 | + | 273 | WP_002362409 | LtrD | - |
| PSC77_RS14135 (PSC77_14135) | 96603..96959 | + | 357 | WP_002393737 | plasmid mobilization relaxosome protein MobC | - |
| PSC77_RS14140 (PSC77_14140) | 96960..98645 | + | 1686 | WP_010784826 | relaxase/mobilization nuclease domain-containing protein | - |
Host bacterium
| ID | 4577 | GenBank | NZ_CP117504 |
| Plasmid name | pJXZ076-99K | Incompatibility group | - |
| Plasmid size | 99966 bp | Coordinate of oriT [Strand] | 96487..96536 [+] |
| Host baterium | Enterococcus faecalis strain CQJXZ21-076 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | prgB/asc10 |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |