Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104122 |
| Name | oriT_2022CK-00781|unnamed5 |
| Organism | Klebsiella grimontii strain 2022CK-00781 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP117735 (29427..29476 [-], 50 nt) |
| oriT length | 50 nt |
| IRs (inverted repeats) | 7..14, 17..24 (GCAAAATT..AATTTTGC) |
| Location of nic site | 33..34 |
| Conserved sequence flanking the nic site |
GGTGTGGTGA |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 50 nt
>oriT_2022CK-00781|unnamed5
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
AAATCTGCAAAATTTTAATTTTGCGTGGGGTGTGGTGATTTTGTGGTGAG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file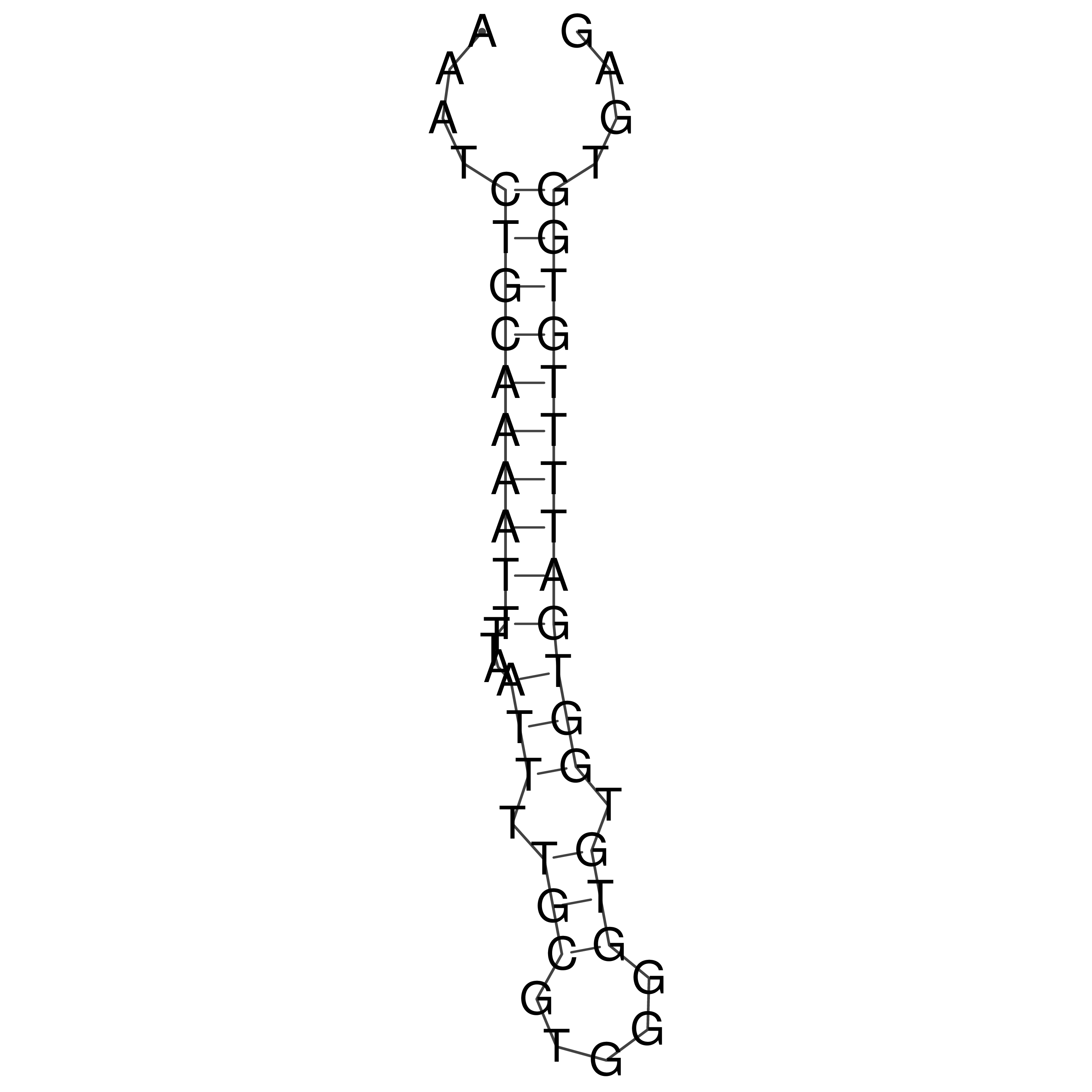
Relaxase
| ID | 3034 | GenBank | WP_273863606 |
| Name | traI_PTC90_RS32135_2022CK-00781|unnamed5 |
UniProt ID | _ |
| Length | 1753 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1753 a.a. Molecular weight: 190608.53 Da Isoelectric Point: 6.2409
>WP_273863606.1 conjugative transfer relaxase/helicase TraI [Klebsiella grimontii]
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAANARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGDRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
MMSIGSVKSAGSAGNYYTDKDNYYVIGSMDERWQGKGAEALGIDGKAVDKALFTELLKGKLPDGSDLTRI
QDGANRHRPGYDLTFSAPKSVSVMAMLGGDKRLIDAHNQAVTEAVRQLETLAATRVMTDGKSETVLTGNL
IVAKFNHDTNRNQEPQIHTHAVVINATQNGDKWQSLGTDKIGKTGFIENVYANQIAFGKLYREAFKPLVE
KLGYETEVVGKHGMWEMKGVPVEPFSTRSQEVREAAGPDASLKSRDVAALDTRKSKEAIDPAEKMVEWMN
TLKETGFDIRGYREAANARAAELARAPAAPVNTDGPDITDVVTKAIAGLSDRKVQFTYADLLARTVGQLE
AKDGVFELARKGIDAAIEREQLIPLDREKGLFTSNIHVLDELAVKALSQEVQRQNHVSVTPDASVVRQVP
FSDAVSVLAQDRPVMGIVSGQGGATGQRERVAELTLMAREQGRDVHILAADNRSRDFLAGDVRLAGETVT
GKSALQDGTAFIPGGTLIVDQAEKLSLKETISLLDGAMRHNVQVLLSDSGKRSGTGSALTVLKDSGVNTY
RWQGGHQTTADIISEPDKGARYSRLAQEFAVSVREGQESVAQISGTREQSVLNGLIRDSLRQEGVLGEKD
TTITALTPVWLDSKSRGVRDYYREGMVMERWDPETRTHDRFVIDRVTASSNMLTLKDRDGDRLDLKVSAV
DSQWTLFRAETLPVAEGERLAVLGKIPDTRLKGGESVTVMKVEEGQLTVQRPGQKTTQTLAVGAGVFDGI
KIGHGWVESPGRSVSETATVFASVTQRELDNATLNQLAQSGSHLRLYSAQDAARTTEKLSRHTAFSVVSE
QLKARSGETDLDAAIAQQKAGLHTPAEQAIHLAIPLLESQDLTFSRPQLLATAMETGGGKVSMADIDTTI
QAQIRSGQLLNVPVAHGYGNDLLISRQTWDAEKSILTHVLEGKGAVAPLMDRVPASLMTDLTAGQRAATR
MILESTDRFTVVQGYAGVGKTTQFRAVMSAISLLPEETRPRVIGLAPTHRAVGEMQSAGVEARTTASFLH
DTQLLQRNGQTPDFSNTLFLLDESSMVGLADMAKAHSLIAAGGGRAVPSGDTDQLPPIASGQPFRLTQQR
SAADVAIMKEIVRQVPELRPAVYSLIERDVHRALTTIEQVTPEQVPRKEGAWAPGSSVVEFTQKQEKEIE
KALSEGKTLPEGQPATLYEALVKDYTGRTPEAQSQTLVITHLNKDRRALNSLIHDARRENGETGKEEITL
PVLVTSNIRDGELRKLSTWTAHKEAVALVDNVYHRISKVDKDNQLITLTDSEGKERFISPREASAEGVTL
YRQEKITVSQGDRMRFSKSDPERGYVANSIWEVQSVSGDSVTLSDGKITRTLTPKAEQAQQHIDLAYAIT
AHGAQGASEPYAIALEGVAGGREQMASFESAYVALSRMKQHVQVYTDNREGWIKAIKNSPEKATAHDILE
PRNDRAVKTADLLFGRARPLDETAAGRAALQQSGLAQGSSPGKFISPGKKYPQPHVALPAFDKNGKAAGI
WLSPLTDRDGRLEAIGGEGRIMGNEDARFVALQNSRNGESLLAGNMGEGVRMARDNPDTGVVVRLAGDDR
PWNPGAMTGGRVWAEPAPVAPVPQAGADIILPPEVLAQRAAEEQQRREMEKQAEQTAREVAGEARKAGEP
ADRVKEVIGDVIRGLERDRPGTEKTTLPDDPQFRRQEAAIQQVASERLQRERLQAVERDMVRDLNREKTL
GGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2924 | GenBank | WP_032072078 |
| Name | traD_PTC90_RS32130_2022CK-00781|unnamed5 |
UniProt ID | _ |
| Length | 770 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 770 a.a. Molecular weight: 85822.78 Da Isoelectric Point: 4.9615
>WP_032072078.1 MULTISPECIES: type IV conjugative transfer system coupling protein TraD [Enterobacteriaceae]
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
MSFNAKDMTQGGQIANMRFRMFGQIANIIFYVLFILFWVLCGLMLMYRLSWQTFVNGCVYWWCTTLGPMR
DIIRSQPVYTIQYYGQSLEYTSEQILADKYTIWCGEQLWTGFAFAAVVSLVICIVTFFIASWVLGRQGKQ
QSEDENTGGRQLSDKPKEVARQMKRDGVASDIKIGDLPILKNSEIQNFCLHGTVGSGKSEVIRRLLNYVR
ARGDMAIIYDRSCEFVKSYYDPSLDKILNPLDSRCAAWDLWKECLTLPDFDNISNTLIPMGTKEDPFWQG
SGRTIFAEGAYLMREDDDRSYEKLVDTMLSIKIDKLRAYLQNTPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEKNGEPFTIRDWMRGVREDRPNGWLFISSNADTHASLKPVISMWLSIAIRGLLAMGENRNRRV
WIFADELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGVKPAATLFDVMNTRAFFRSPSKEIA
EFAAGEIGEKEILKASEQYSYGADPVRDGVSTGKEKERETLVSYSDIQTLPDLSCYVTLPGPYPAVKLAL
KYKPRPKIAEGFIPRTLDARVDARLSALLEAREAEGSLARALFTPDAPASGPADTNSHAGEQPEPVSQPA
PADMTVSPAPVKAPPTTKMPAEEPSVRATEPSVLRLTTVPLIKPKAAAAAAAAATASSAGTPAAAAGGTE
QELAQQSAEQGQDMLPAGMNEDGVIEDMQAYDAWLADEQTQRDMQRREEVNINHSHRHDEQDDVEIGGNF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 31935..59078
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PTC90_RS31950 (PTC90_31945) | 28295..28837 | + | 543 | WP_020804853 | antirestriction protein | - |
| PTC90_RS31955 (PTC90_31950) | 28860..29282 | - | 423 | WP_020315406 | transglycosylase SLT domain-containing protein | - |
| PTC90_RS31960 (PTC90_31955) | 29776..30174 | + | 399 | WP_004152493 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PTC90_RS31965 (PTC90_31960) | 30348..31034 | + | 687 | WP_004152494 | transcriptional regulator TraJ family protein | - |
| PTC90_RS31970 (PTC90_31965) | 31113..31499 | + | 387 | WP_004152495 | TraY domain-containing protein | - |
| PTC90_RS31975 (PTC90_31970) | 31553..31921 | + | 369 | WP_004194426 | type IV conjugative transfer system pilin TraA | - |
| PTC90_RS31980 (PTC90_31975) | 31935..32240 | + | 306 | WP_004178059 | type IV conjugative transfer system protein TraL | traL |
| PTC90_RS31985 (PTC90_31980) | 32260..32826 | + | 567 | WP_115808217 | type IV conjugative transfer system protein TraE | traE |
| PTC90_RS31990 (PTC90_31985) | 32813..33553 | + | 741 | WP_013214019 | type-F conjugative transfer system secretin TraK | traK |
| PTC90_RS31995 (PTC90_31990) | 33553..34977 | + | 1425 | WP_032426793 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PTC90_RS32000 (PTC90_31995) | 35091..35315 | + | 225 | Protein_50 | type IV conjugative transfer system lipoprotein TraV | - |
| PTC90_RS32005 (PTC90_32000) | 35866..36138 | + | 273 | WP_273863597 | hypothetical protein | - |
| PTC90_RS32010 (PTC90_32005) | 36229..36521 | + | 293 | Protein_52 | hypothetical protein | - |
| PTC90_RS32015 (PTC90_32010) | 36545..36777 | + | 233 | Protein_53 | hypothetical protein | - |
| PTC90_RS32020 (PTC90_32015) | 36809..37052 | + | 244 | Protein_54 | hypothetical protein | - |
| PTC90_RS32025 (PTC90_32020) | 37343..37709 | + | 367 | Protein_55 | hypothetical protein | - |
| PTC90_RS32030 (PTC90_32025) | 37778..40411 | + | 2634 | Protein_56 | type IV secretion system protein TraC | - |
| PTC90_RS32035 (PTC90_32030) | 40510..40844 | + | 335 | Protein_57 | type-F conjugative transfer system protein TrbI | - |
| PTC90_RS32040 (PTC90_32035) | 40844..41470 | + | 627 | WP_020314628 | type-F conjugative transfer system protein TraW | traW |
| PTC90_RS32045 (PTC90_32040) | 41514..42473 | + | 960 | WP_029497356 | conjugal transfer pilus assembly protein TraU | traU |
| PTC90_RS32050 (PTC90_32045) | 42488..43036 | + | 549 | WP_022631518 | hypothetical protein | - |
| PTC90_RS32055 (PTC90_32050) | 43757..44359 | + | 603 | WP_022631520 | hypothetical protein | - |
| PTC90_RS32060 (PTC90_32055) | 44504..45151 | + | 648 | WP_039698503 | type-F conjugative transfer system pilin assembly protein TrbC | trbC |
| PTC90_RS32065 (PTC90_32060) | 45210..47165 | + | 1956 | WP_039698502 | type-F conjugative transfer system mating-pair stabilization protein TraN | traN |
| PTC90_RS32070 (PTC90_32065) | 47198..47479 | + | 282 | WP_032419932 | hypothetical protein | - |
| PTC90_RS32075 (PTC90_32070) | 47469..47696 | + | 228 | WP_012540032 | conjugal transfer protein TrbE | - |
| PTC90_RS32080 (PTC90_32075) | 47707..48033 | + | 327 | WP_273863599 | hypothetical protein | - |
| PTC90_RS32085 (PTC90_32080) | 48054..48806 | + | 753 | WP_004152677 | type-F conjugative transfer system pilin assembly protein TraF | traF |
| PTC90_RS32090 (PTC90_32085) | 48817..49056 | + | 240 | WP_004144400 | type-F conjugative transfer system pilin chaperone TraQ | - |
| PTC90_RS32095 (PTC90_32090) | 49028..49585 | + | 558 | WP_013214031 | type-F conjugative transfer system pilin assembly thiol-disulfide isomerase TrbB | traF |
| PTC90_RS32100 (PTC90_32095) | 49631..50074 | + | 444 | WP_004194305 | F-type conjugal transfer protein TrbF | - |
| PTC90_RS32105 (PTC90_32100) | 50052..51431 | + | 1380 | WP_273863600 | conjugal transfer pilus assembly protein TraH | traH |
| PTC90_RS32110 (PTC90_32105) | 51431..54280 | + | 2850 | WP_273863602 | conjugal transfer mating-pair stabilization protein TraG | traG |
| PTC90_RS32115 (PTC90_32110) | 54293..54841 | + | 549 | WP_004152623 | conjugal transfer entry exclusion protein TraS | - |
| PTC90_RS32120 (PTC90_32115) | 55025..55756 | + | 732 | WP_004152622 | conjugal transfer complement resistance protein TraT | - |
| PTC90_RS32125 (PTC90_32120) | 55948..56637 | + | 690 | WP_022644928 | hypothetical protein | - |
| PTC90_RS32130 (PTC90_32125) | 56766..59078 | + | 2313 | WP_032072078 | type IV conjugative transfer system coupling protein TraD | virb4 |
Host bacterium
| ID | 4561 | GenBank | NZ_CP117735 |
| Plasmid name | 2022CK-00781|unnamed5 | Incompatibility group | - |
| Plasmid size | 69503 bp | Coordinate of oriT [Strand] | 29427..29476 [-] |
| Host baterium | Klebsiella grimontii strain 2022CK-00781 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |