Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104082 |
| Name | oriT_pEA25_1 |
| Organism | Escherichia albertii strain BIA_25 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP117616 (44017..44102 [-], 86 nt) |
| oriT length | 86 nt |
| IRs (inverted repeats) | 61..68, 73..80 (TTGGTGGT..ACCACCAA) 27..34, 37..44 (GCAAAAAC..GTTTTTGC) 8..13, 21..26 (TGATTT..AAATCA) |
| Location of nic site | 53..54 |
| Conserved sequence flanking the nic site |
GGTGTGGTGC |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 86 nt
AATTACATGATTTAAAACGCAAATCAGCAAAAACTTGTTTTTGCGTGGGGTGTGGTGCTTTTGGTGGTGAGAACCACCAACCTGTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file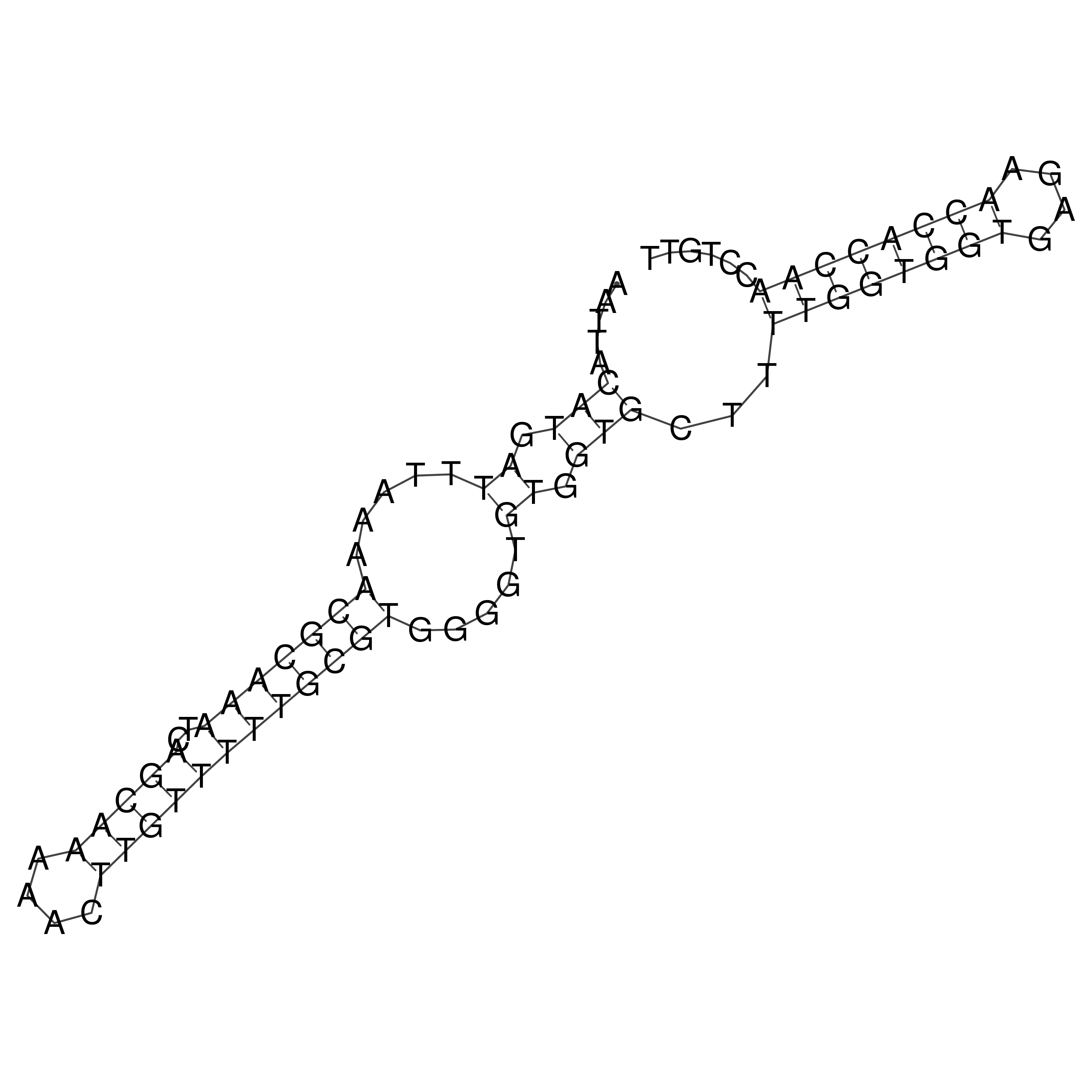
Relaxase
| ID | 3018 | GenBank | WP_273813309 |
| Name | traI_PS046_RS23600_pEA25_1 |
UniProt ID | _ |
| Length | 1756 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1756 a.a. Molecular weight: 192496.10 Da Isoelectric Point: 6.0524
MMSIAQVRSAGSAGNYYTDKDNYYVLGSMGERWAGQGAEQLGLQGSVDKDLFTRLLEGRLPDGADLSRMQ
DGRNRHRPGYDLTFSAPKSVSMMAMPGGDKRLIDAHNQAVDFAVRQVEALASTRVMTDGQSETVLTGNLV
MALFNHDTSRDQEPQLHTHVVVANVTQHNGEWKTLSSDKVGKTGFIENVYANQITFGRLYREKLKEQVEA
LGYETEVVGKHGMWEMPGVPVEAFSGRSQTIREAVGEDASLKSRDVAALDTRKSKQHVDPEVRMTEWLQT
LKETGFDIRAYRDAADQRAEIRTQTPGPASQDWPDVQQAVTQAIAGLSERKVQFTYTDVLARTVGILPPG
NGVIDRARAGIDEAISREQLIPLDREKGLFTSGIHVLDELSVRALSRDIMKQNRVAVHPEKSVPRTACYS
DAVSVLAQDRPSLAIVSGQGGAAGQRERVAELVMMAREQGREVQIIAADRRSQMNLKQDERLSGELITGR
RQLQEGMAFTPGNTVIVDQGEKLSLKETLTLLDGAARHNVQVLITDSGQRTGTGSALMAMKDAGVNTYRW
QGGEQRPATIISEPDRNVRYARLAGDFAASVKAGEESVAQVSGVREQAILTQAIRSELKTQGVLGHQEVT
MTALSPVWLDSRSRYLRDMYRPGMVMEQWNPETRSHDRYVIDRVTAQSHSLTLRDAQGETQVVRISSLDS
SWSLFRPEKMPVADGERLRVTGKIPGLRVYGGDRLQAASVSEDAMTVVVPGRAEPATLPVADSPFTALKL
ENGWVETPGHSVSDSAKVFASVTQMAMDNATLNGLARSGRDVRLYSSLDEPRTAEKLARHPSFTVVSEQI
KARAGETSLETAISHQKSALHTPAQQAIHLALPVVESKNLAFSHVDLLTEAKSFAAEGTSFTELGREIDA
QIKRGDLLHVDVAKGYGTGLLVSRASYEAEKSILRHILEGKEAVTPLMERVPGELMEKLTSGQRAATRMI
LETSDRFTVVQGYAGVGKTTQFRAVMSAVNMLPESERPRVVGLGPTHRAVGEMRSAGVDAQTLASFLHDT
QLQQRSGETPDFSNTLFLLDESSMVGNTDMARAYALIAAGGGRAVASGDTDQLQAIAPGQPFRLQQTRSA
ADVAIMKEIVRQTPELREAVYSLINRDVERALSGLESVKPSQVPRLEGAWAPERSVTEFSHSQEAKLAEA
QQKAMLKGEAFPDVPMTLYEAIVRDYTGRTPEAREQTLIVTHLNEDRRVLNSMIHDAREKAGELGKEQVM
VPVLNTANIRDGELRRLSTWETHRDALVLVDNVYHRIAGISKDDGLITLQDAEGNTRLISPREAVAEGVT
LYTPDTIRVGTGDRMRFTKSDRERGYVANSVWTVTAVSGDSVTLSDGQQTRVIHPGQERAEQHIDLAYAI
TAHGAQGASETFAIALEGTEGNRKQMAGFESAYVALSRMKQHVQVYTDNRQGWTDAINNAVQKGTAHDVF
EPKPDREVMNAERLFSTARELRDVAAGRAVLRQAGLAGGDSPARFIAPGRKYPQPYVALPAFDRNGKSAG
IWLNPLTTDDGNGLRGFSGEGRVKGSGDAQFVALQGSRNGESLLADNMQDGVRIARDNPDSGVVVRIAGE
GRPWNPGAITGGRVWGDIPDNSVQPGAGNGEPVTAEVLAQRQAEEAIRRETERRADEIVRKMAEEKPDHP
DDKTEQAVREIAGQERDRAAISEREAALPESVLREPQREREAVREVARENLLQERLQQMERDMVRDLQKE
KTLGGD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Auxiliary protein
| ID | 1282 | GenBank | WP_161527390 |
| Name | WP_161527390_pEA25_1 |
UniProt ID | _ |
| Length | 127 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 127 a.a. Molecular weight: 14536.52 Da Isoelectric Point: 4.7116
MARVILYISNDVYDKVNAIVEQRRQEGARDKDISLSGTASMLLELGLRVYEAQMERKESAFNQTEFNKLL
LECVVKTQSTVAKILGIESLSPHVSGNPKFEYANMVEDIREKVSSEMERFFPKNDEE
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 1283 | GenBank | WP_072247434 |
| Name | WP_072247434_pEA25_1 |
UniProt ID | _ |
| Length | 75 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
Auxiliary protein sequence
Download Length: 75 a.a. Molecular weight: 8556.84 Da Isoelectric Point: 10.7978
MSRNIIRPAPGNKVLLVLDDATNHKLLGARERSGRTKTNEVLVRLRDHLRRFPDFYNIDSIKEGTGKTDS
IIKDL
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2897 | GenBank | WP_273813308 |
| Name | traD_PS046_RS23585_pEA25_1 |
UniProt ID | _ |
| Length | 729 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 729 a.a. Molecular weight: 82932.69 Da Isoelectric Point: 5.1048
MSFNAKDMTQGGQIASMRIRMFSQIANIMLYCLFIFFWILVGLVLWVKISWQTFVNGCIYWWCTTLEGMR
DLIKSQPVYEIQYYGKTFRMNAAQVLHDKYMIWCGEQLWSAFVLATVVALVICLITFFVVSWILGRQGKQ
QSENEVTGGRQLTDNPKDVARMLKKDGKDSDIRIGDLPIIRDSEIQNFCLHGTVGAGKSEVIRRLANYAR
QRGDMVVIYDRSGEFVKSYYDPSIDKILNPLDARCAAWDLWKECLTQPDFDNTANTLIPMGTKEDPFWQG
SGRTIFAEAAYLMRNDPNRSYSKLVDTLLSIKIEKLRTFLRNSPAANLVEEKIEKTAISIRAVLTNYVKA
IRYLQGIEHNGEPFTIRDWMRGVREDQKNGWLFISSNADTHASLKPVISMWLSIAIRGLMAMGENRNRRV
WFFCDELPTLHKLPDLVEILPEARKFGGCYVFGIQSYAQLEDIYGEKSAATLFDVMNTRAFFRSPSHKIA
EFAAGEIGEKEHLKASLQYSYGADPVRDGVSTGKDMERQTLVSYSDIQSLPDLTCYVTLPGPYPAVKLSL
KYQARPKVAPEFIPREINPEMENRLSAVLAAREAEGRQMASLFEPDMPEVVSGEDVTQAEQPQQPQQPQQ
PQQPQQPVSPVINDKKSDAGVNVPAGGIEQELKMKPEEEMEQQLPPGISESGEVVDMAAYEAWQQENHPD
IQQQMQRREEVNINVHRERGEDVEPGDDF
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 43449..59362
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| PS046_RS23460 (PS046_23460) | 39383..39946 | + | 564 | WP_021548773 | class I SAM-dependent methyltransferase | - |
| PS046_RS23465 (PS046_23465) | 40035..40457 | - | 423 | Protein_43 | hypothetical protein | - |
| PS046_RS23470 (PS046_23470) | 40736..41740 | - | 1005 | WP_273813360 | IS110 family transposase | - |
| PS046_RS23475 (PS046_23475) | 41924..42211 | + | 288 | WP_085457081 | hypothetical protein | - |
| PS046_RS23480 (PS046_23480) | 42332..43153 | + | 822 | WP_032342235 | DUF932 domain-containing protein | - |
| PS046_RS23485 (PS046_23485) | 43449..44051 | - | 603 | WP_207610769 | transglycosylase SLT domain-containing protein | virB1 |
| PS046_RS23490 (PS046_23490) | 44374..44757 | + | 384 | WP_161527390 | conjugal transfer relaxosome DNA-binding protein TraM | - |
| PS046_RS23495 (PS046_23495) | 44952..45623 | + | 672 | WP_059257109 | conjugal transfer transcriptional regulator TraJ | - |
| PS046_RS23500 (PS046_23500) | 45761..45988 | + | 228 | WP_072247434 | conjugal transfer relaxosome protein TraY | - |
| PS046_RS23505 (PS046_23505) | 46021..46380 | + | 360 | WP_059257108 | type IV conjugative transfer system pilin TraA | - |
| PS046_RS23510 (PS046_23510) | 46395..46706 | + | 312 | WP_000012118 | type IV conjugative transfer system protein TraL | traL |
| PS046_RS23515 (PS046_23515) | 46728..47294 | + | 567 | WP_059259957 | type IV conjugative transfer system protein TraE | traE |
| PS046_RS23520 (PS046_23520) | 47307..48008 | + | 702 | WP_273813362 | type-F conjugative transfer system secretin TraK | traK |
| PS046_RS23525 (PS046_23525) | 48008..49435 | + | 1428 | WP_273813352 | F-type conjugal transfer pilus assembly protein TraB | traB |
| PS046_RS23530 (PS046_23530) | 49425..50009 | + | 585 | WP_273813354 | conjugal transfer pilus-stabilizing protein TraP | - |
| PS046_RS23535 (PS046_23535) | 49996..50316 | + | 321 | WP_273813356 | conjugal transfer protein TrbD | - |
| PS046_RS23540 (PS046_23540) | 50313..50825 | + | 513 | WP_161527393 | type IV conjugative transfer system lipoprotein TraV | traV |
| PS046_RS23545 (PS046_23545) | 50960..51022 | + | 63 | Protein_59 | conjugal transfer protein TraR | - |
| PS046_RS23550 (PS046_23550) | 51076..51753 | + | 678 | WP_001339397 | IS66-like element accessory protein TnpA | - |
| PS046_RS23555 (PS046_23555) | 51753..52100 | + | 348 | WP_000624622 | IS66 family insertion sequence element accessory protein TnpB | - |
| PS046_RS23560 (PS046_23560) | 52120..53691 | + | 1572 | WP_273811869 | IS66 family transposase | - |
| PS046_RS23565 (PS046_23565) | 53722..54213 | + | 492 | Protein_63 | complement resistance protein TraT | - |
| PS046_RS23570 (PS046_23570) | 54481..56052 | - | 1572 | WP_273811869 | IS66 family transposase | - |
| PS046_RS23575 (PS046_23575) | 56072..56419 | - | 348 | WP_273813306 | IS66 family insertion sequence element accessory protein TnpB | - |
| PS046_RS23580 (PS046_23580) | 56419..57096 | - | 678 | WP_001339397 | IS66-like element accessory protein TnpA | - |
| PS046_RS23585 (PS046_23585) | 57173..59362 | + | 2190 | WP_273813308 | type IV conjugative transfer system coupling protein TraD | virb4 |
| PS046_RS23590 (PS046_23590) | 59371..59769 | - | 399 | WP_059257082 | tRNA(fMet)-specific endonuclease VapC | - |
| PS046_RS23595 (PS046_23595) | 59769..59996 | - | 228 | WP_000450526 | toxin-antitoxin system antitoxin VapB | - |
Host bacterium
| ID | 4521 | GenBank | NZ_CP117616 |
| Plasmid name | pEA25_1 | Incompatibility group | IncFIB |
| Plasmid size | 100741 bp | Coordinate of oriT [Strand] | 44017..44102 [-] |
| Host baterium | Escherichia albertii strain BIA_25 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | gspG, gspH, vat |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |