Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104036 |
| Name | oriT_pSymA |
| Organism | Sinorhizobium meliloti strain AK76 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP066361 (856213..856241 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pSymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file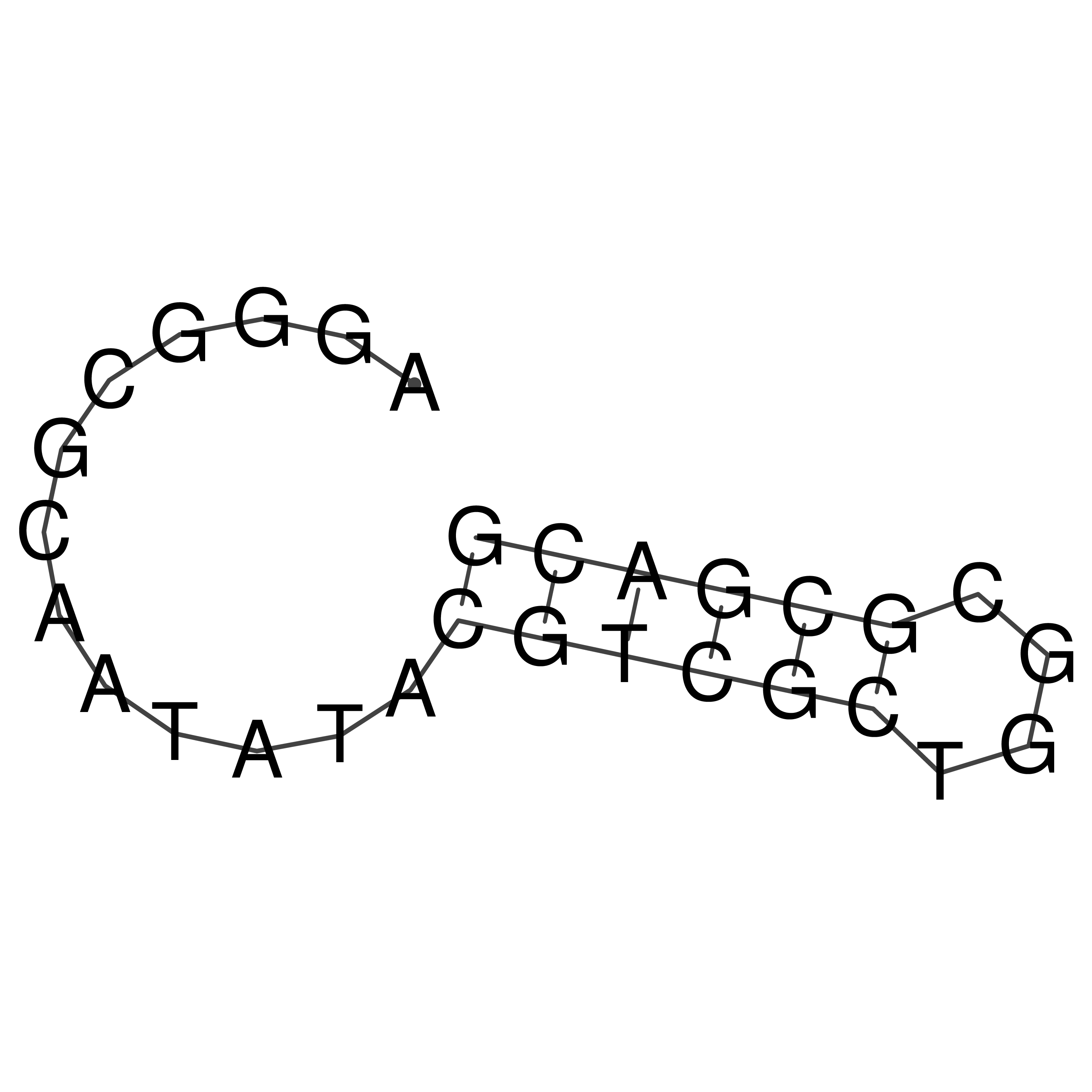
Relaxase
| ID | 2991 | GenBank | WP_198868964 |
| Name | traA_JFX10_RS29660_pSymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170140.81 Da Isoelectric Point: 9.4868
>WP_198868964.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQIDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAKIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWDRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSIPALSKTAMARLVEIEAVRKQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDRKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
QSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEQGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVAMYFAPADLARRQEMADRLLAEPELLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMASDDLVLLKPQQIDAETGEAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARAAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGREQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQHGRIVETETRAKIVDRIVADWADARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISQDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWDRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSIPALSKTAMARLVEIEAVRKQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDRKLDVAAGSLDPRIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRVQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2857 | GenBank | WP_198868965 |
| Name | traG_JFX10_RS29675_pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70228.22 Da Isoelectric Point: 9.4769
>WP_198868965.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKTHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
MALKAKTHPSLLVILFPVAVTAAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWVL
PLHRRRPVAIASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAVP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 774451..784029
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| JFX10_RS29230 (JFX10_29215) | 771333..772181 | + | 849 | WP_017269096 | alpha/beta fold hydrolase | - |
| JFX10_RS29235 (JFX10_29220) | 772231..772878 | + | 648 | WP_010967700 | dihydrofolate reductase family protein | - |
| JFX10_RS29240 (JFX10_29225) | 772952..773203 | + | 252 | WP_017269095 | hypothetical protein | - |
| JFX10_RS29245 (JFX10_29230) | 773464..773832 | - | 369 | WP_013845467 | hypothetical protein | - |
| JFX10_RS33715 | 773929..774447 | + | 519 | WP_013845466 | hypothetical protein | - |
| JFX10_RS29250 (JFX10_29235) | 774451..775122 | + | 672 | WP_198869109 | lytic transglycosylase domain-containing protein | virB1 |
| JFX10_RS29255 (JFX10_29240) | 775119..775418 | + | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| JFX10_RS29260 (JFX10_29245) | 775423..775764 | + | 342 | WP_003526740 | type IV secretion system protein VirB3 | virB3 |
| JFX10_RS29265 (JFX10_29250) | 775739..778135 | + | 2397 | WP_198869110 | VirB4 family type IV secretion system protein | virb4 |
| JFX10_RS29270 (JFX10_29255) | 778135..778836 | + | 702 | WP_014531544 | P-type DNA transfer protein VirB5 | virB5 |
| JFX10_RS29275 (JFX10_29260) | 778833..779048 | + | 216 | WP_014531545 | EexN family lipoprotein | - |
| JFX10_RS29280 (JFX10_29265) | 779071..780006 | + | 936 | WP_014531546 | type IV secretion system protein | virB6 |
| JFX10_RS33720 | 780003..780317 | + | 315 | WP_013845458 | hypothetical protein | - |
| JFX10_RS29285 (JFX10_29270) | 780322..780993 | + | 672 | WP_127514435 | virB8 family protein | virB8 |
| JFX10_RS29290 (JFX10_29275) | 780993..781847 | + | 855 | WP_014531548 | P-type conjugative transfer protein VirB9 | virB9 |
| JFX10_RS29295 (JFX10_29280) | 781857..783029 | + | 1173 | WP_198869111 | type IV secretion system protein VirB10 | virB10 |
| JFX10_RS29300 (JFX10_29285) | 783040..784029 | + | 990 | WP_198869112 | P-type DNA transfer ATPase VirB11 | virB11 |
| JFX10_RS29305 (JFX10_29290) | 784340..785566 | - | 1227 | WP_013845453 | MFS transporter | - |
| JFX10_RS29310 (JFX10_29295) | 785603..786214 | - | 612 | WP_013845452 | TetR/AcrR family transcriptional regulator | - |
| JFX10_RS29315 (JFX10_29300) | 786741..786926 | + | 186 | WP_013845451 | periplasmic nitrate reductase, NapE protein | - |
| JFX10_RS29320 (JFX10_29305) | 786926..787426 | + | 501 | WP_013845450 | ferredoxin-type protein NapF | - |
| JFX10_RS29325 (JFX10_29310) | 787419..787706 | + | 288 | WP_013845449 | chaperone NapD | - |
Host bacterium
| ID | 4475 | GenBank | NZ_CP066361 |
| Plasmid name | pSymA | Incompatibility group | - |
| Plasmid size | 1580177 bp | Coordinate of oriT [Strand] | 856213..856241 [-] |
| Host baterium | Sinorhizobium meliloti strain AK76 |
Cargo genes
| Drug resistance gene | aac(3)-IIc |
| Virulence gene | htpB, katA |
| Metal resistance gene | ncrA |
| Degradation gene | - |
| Symbiosis gene | fixA, fixB, fixN, fixO, fixG, fixS, nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixC, fixX, nifB, fixU, nifT |
| Anti-CRISPR | - |