Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 104010 |
| Name | oriT_pA |
| Organism | Sinorhizobium meliloti WSM1022 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP054392 (947549..947577 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file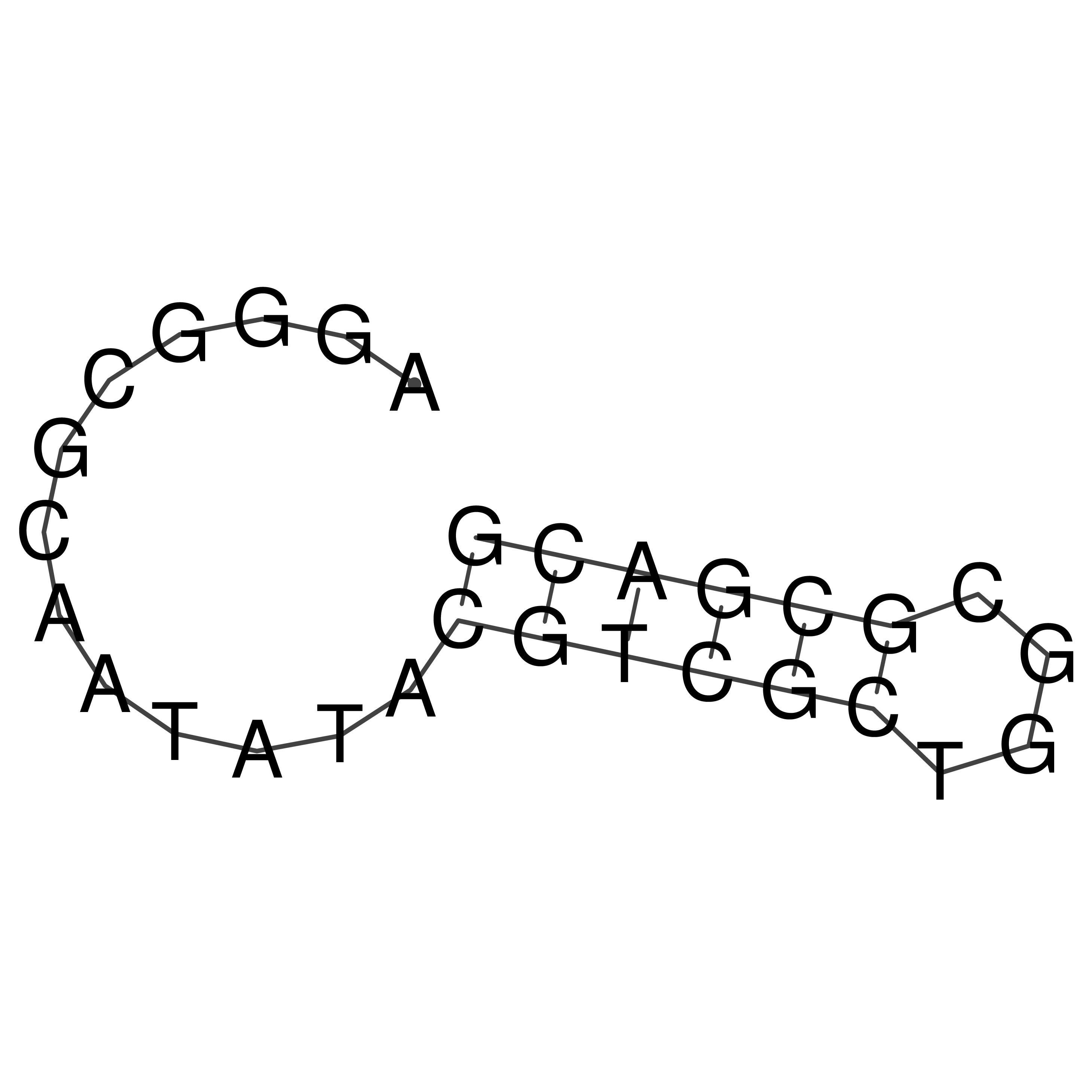
Relaxase
| ID | 2973 | GenBank | WP_174167964 |
| Name | traA_HR059_RS29635_pA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170141.90 Da Isoelectric Point: 9.3645
>WP_174167964.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMTSDELVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPLSGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMTLSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLRAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVITPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMTSDELVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPLSGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMTLSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAF
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAPVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2834 | GenBank | WP_028011681 |
| Name | virD4_HR059_RS26780_pA |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 62923.22 Da Isoelectric Point: 9.1751
>WP_028011681.1 type IV secretion system ATPase VirD4 [Sinorhizobium meliloti]
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILLVQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDILALPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPAPLMIADERF
SHV
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILLVQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDILALPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPAPLMIADERF
SHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2835 | GenBank | WP_028011664 |
| Name | traG_HR059_RS29650_pA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70413.41 Da Isoelectric Point: 9.3555
>WP_028011664.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLLILLPVAVTAFAIYVVGWRWPGLAAGMSGKTAYWFLRASPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGLLSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPDDPATWGKGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSSGAYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSQSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSTDIVTGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLLILLPVAVTAFAIYVVGWRWPGLAAGMSGKTAYWFLRASPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGLLSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPDDPATWGKGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSSGAYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSQSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSTDIVTGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 335426..344839
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HR059_RS26685 (HR059_26685) | 330507..331964 | + | 1458 | WP_028011837 | PLP-dependent aminotransferase family protein | - |
| HR059_RS26690 (HR059_26690) | 332291..332562 | - | 272 | Protein_320 | hypothetical protein | - |
| HR059_RS26695 (HR059_26695) | 332612..333808 | - | 1197 | WP_174167946 | IS256-like element ISRm5 family transposase | - |
| HR059_RS26700 (HR059_26700) | 333900..334466 | - | 567 | WP_028011686 | type IV secretory system conjugative DNA transfer family protein | - |
| HR059_RS26705 (HR059_26705) | 334450..334641 | + | 192 | Protein_323 | Lrp/AsnC ligand binding domain-containing protein | - |
| HR059_RS26710 (HR059_26710) | 334700..335098 | + | 399 | WP_164820336 | hypothetical protein | - |
| HR059_RS26715 (HR059_26715) | 335426..336133 | + | 708 | WP_028011685 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| HR059_RS26720 (HR059_26720) | 336147..336509 | + | 363 | WP_014528518 | pilin major subunit VirB2 | virB2 |
| HR059_RS26725 (HR059_26725) | 336506..336832 | + | 327 | WP_003537077 | type IV secretion system protein VirB3 | virB3 |
| HR059_RS26730 (HR059_26730) | 336832..339201 | + | 2370 | WP_028011684 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| HR059_RS26735 (HR059_26735) | 339215..339898 | + | 684 | WP_003537081 | pilin minor subunit VirB5 | traI |
| HR059_RS26740 (HR059_26740) | 339979..340866 | + | 888 | WP_003537089 | type IV secretion system protein | virB6 |
| HR059_RS26745 (HR059_26745) | 340899..341066 | + | 168 | WP_003537091 | type IV secretion system lipoprotein VirB7 | - |
| HR059_RS26750 (HR059_26750) | 341053..341766 | + | 714 | WP_003537093 | type IV secretion system protein VirB8 | virB8 |
| HR059_RS26755 (HR059_26755) | 341763..342644 | + | 882 | WP_028011683 | P-type conjugative transfer protein VirB9 | virB9 |
| HR059_RS26760 (HR059_26760) | 342641..343759 | + | 1119 | WP_028011682 | type IV secretion system protein VirB10 | virB10 |
| HR059_RS26765 (HR059_26765) | 343799..344839 | + | 1041 | WP_017263499 | P-type DNA transfer ATPase VirB11 | virB11 |
| HR059_RS26770 (HR059_26770) | 346077..346802 | + | 726 | WP_017263500 | response regulator | - |
| HR059_RS26775 (HR059_26775) | 347001..347177 | + | 177 | WP_153323329 | hypothetical protein | - |
| HR059_RS26780 (HR059_26780) | 347219..348910 | - | 1692 | WP_028011681 | type IV secretion system ATPase VirD4 | - |
Region 2: 822940..832519
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| HR059_RS28995 (HR059_28995) | 819827..820675 | + | 849 | WP_017264156 | alpha/beta fold hydrolase | - |
| HR059_RS29000 (HR059_29000) | 820725..821372 | + | 648 | WP_017264157 | dihydrofolate reductase family protein | - |
| HR059_RS29005 (HR059_29005) | 821491..821694 | + | 204 | WP_033052013 | hypothetical protein | - |
| HR059_RS29010 (HR059_29010) | 821953..822321 | - | 369 | WP_017264159 | transcriptional regulator | - |
| HR059_RS32185 | 822418..822936 | + | 519 | WP_017264160 | hypothetical protein | - |
| HR059_RS29015 (HR059_29015) | 822940..823611 | + | 672 | WP_017264161 | lytic transglycosylase domain-containing protein | virB1 |
| HR059_RS29020 (HR059_29020) | 823608..823907 | + | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| HR059_RS29025 (HR059_29025) | 823912..824253 | + | 342 | WP_017264162 | type IV secretion system protein VirB3 | virB3 |
| HR059_RS29030 (HR059_29030) | 824228..826624 | + | 2397 | WP_174167962 | VirB4 family type IV secretion system protein | virb4 |
| HR059_RS29035 (HR059_29035) | 826624..827325 | + | 702 | WP_010967692 | P-type DNA transfer protein VirB5 | virB5 |
| HR059_RS29040 (HR059_29040) | 827322..827555 | + | 234 | WP_013845460 | EexN family lipoprotein | - |
| HR059_RS29045 (HR059_29045) | 827561..828496 | + | 936 | WP_013845459 | type IV secretion system protein | virB6 |
| HR059_RS29050 (HR059_29050) | 828493..828807 | + | 315 | WP_028010966 | hypothetical protein | - |
| HR059_RS29055 (HR059_29055) | 828812..829483 | + | 672 | WP_010967689 | virB8 family protein | virB8 |
| HR059_RS29060 (HR059_29060) | 829483..830337 | + | 855 | WP_014528658 | P-type conjugative transfer protein VirB9 | virB9 |
| HR059_RS29065 (HR059_29065) | 830347..831519 | + | 1173 | WP_010967687 | type IV secretion system protein VirB10 | virB10 |
| HR059_RS29070 (HR059_29070) | 831530..832519 | + | 990 | WP_010967686 | P-type DNA transfer ATPase VirB11 | virB11 |
| HR059_RS29075 (HR059_29075) | 832831..834057 | - | 1227 | WP_010967685 | MFS transporter | - |
| HR059_RS29080 (HR059_29080) | 834092..834703 | - | 612 | WP_010967684 | TetR/AcrR family transcriptional regulator | - |
| HR059_RS29085 (HR059_29085) | 834955..835632 | + | 678 | WP_010967683 | YoaK family protein | - |
| HR059_RS29090 (HR059_29090) | 835879..836901 | + | 1023 | WP_028010967 | alcohol dehydrogenase AdhP | - |
Host bacterium
| ID | 4450 | GenBank | NZ_CP054392 |
| Plasmid name | pA | Incompatibility group | - |
| Plasmid size | 1383549 bp | Coordinate of oriT [Strand] | 947549..947577 [-] |
| Host baterium | Sinorhizobium meliloti WSM1022 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixN, fixO, fixG, fixS, nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifB, fixU |
| Anti-CRISPR | - |