Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103926 |
| Name | oriT_pAtCFBP7129a |
| Organism | Agrobacterium tumefaciens strain CFBP7129 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP039924 (361545..361598 [-], 54 nt) |
| oriT length | 54 nt |
| IRs (inverted repeats) | 23..29, 34..40 (ACGTCGC..GCGACGT) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 54 nt
>oriT_pAtCFBP7129a
CCCCGCCAGGGCTGCAATCGGGACGTCGCGGCAGCGACGTATAATTGCGCCCTT
CCCCGCCAGGGCTGCAATCGGGACGTCGCGGCAGCGACGTATAATTGCGCCCTT
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file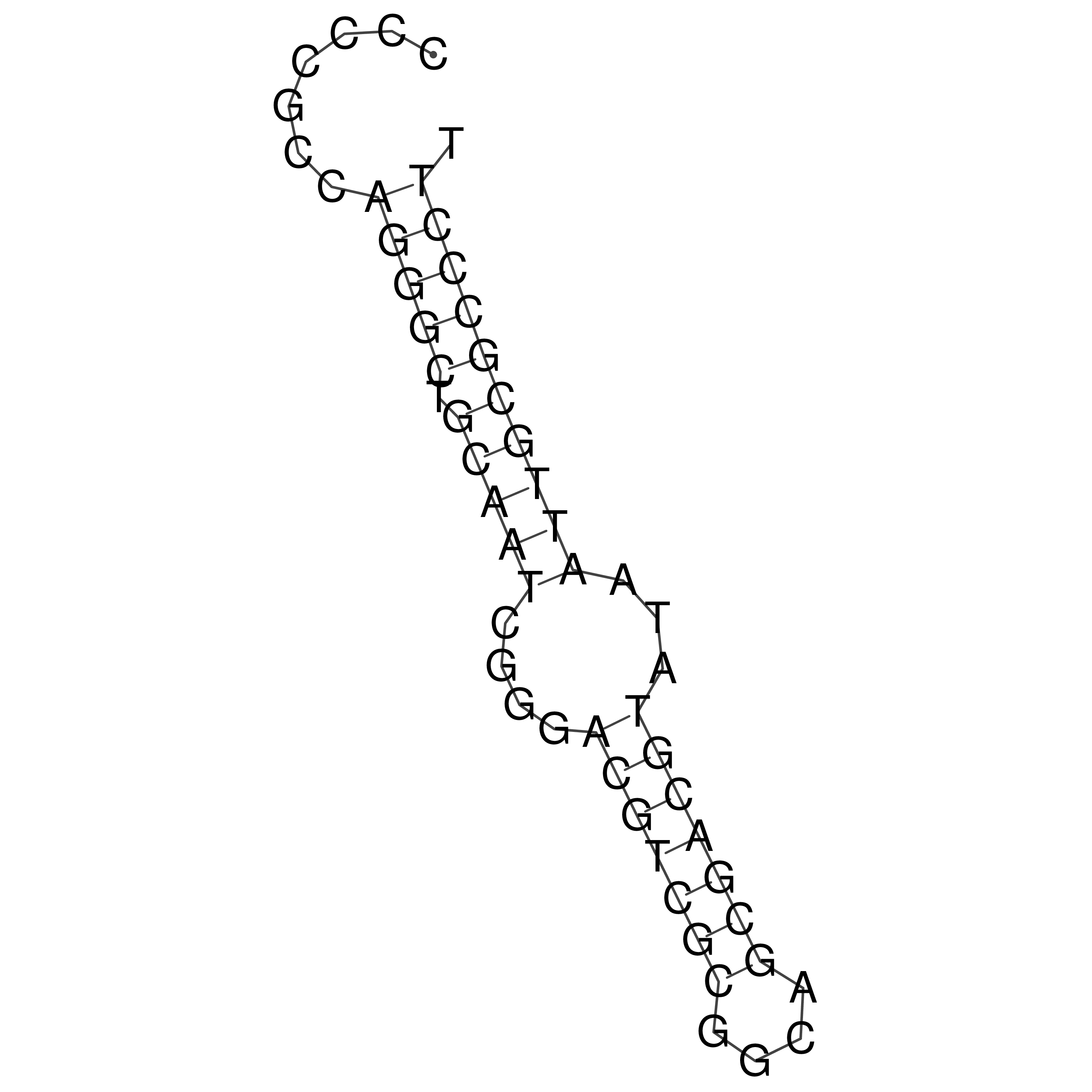
Relaxase
| ID | 2918 | GenBank | WP_137006245 |
| Name | traA_CFBP7129_RS27430_pAtCFBP7129a |
UniProt ID | _ |
| Length | 1108 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1108 a.a. Molecular weight: 123232.24 Da Isoelectric Point: 10.0046
>WP_137006245.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARVIDYTRKQDLLHEEFVVPADTPEWLRSMISAR
SVALRSEAFWNKVEEFEKRADAQLARDVTIALPIELTAGQNIALVRDFVERHVTAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKRIAVTGPDGKPMRNDAGKIVYELWAGSVDDFNAFRDGWFACQNHHLALAG
FDVRIDGRSFEKQAVDLEPTIHLGVGTKAIERKREQSGRKREAFPSRLERIEIQDARRVENTRRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDGRLFQSLMVRILQSPQALRLERERINLATGTRSPAKYTTREM
IRLEAGMASRAIWLSGRASHGVPGTVIEATFARHPGLSREQQTAIEHAAGAGRIAVVIGRAGAGKTTMMK
AGREAWEGAGYRVVGGALAGKATEGLEKEAGIQSRTLSSWELRWKQGRDQLDKKTVFILDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRTGYAELETIYRQREQWMRDASLDLARGSVGKA
VDAYTAHGRMIGSRLKDEAVESLIAAWDRDHDPSKTSLILAHLRRDVRMLNDMARARLVERGVVADGFSF
TTEDGVRMFAAGDQIVFLRNEGSLGVRNGMLARVLEAVPGRIVAETGEGEFRRRVTVEQRFYNNLDHGYA
TTIHKAQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGNRSFAKAGGLVPILSRQNAKETTLDYE
NASFYRQALRFAETRGLHLVNVARTIARDRLEWTLRQKQKLVDLGARLAAIGRAPGLLRGSKNQSIPDPI
KEARPMVSGITIFPKSLEQAVEDKLAGDPGLKKQWQDVSSRFHLVYAQPEAAFNAINVDAMIIDQSVAKS
TLARIAGEPESFGALNGKTGLLASRMEKQKRETAIANVPALARNLERYLRERAEAELKHEREERAARLKV
SIDIPALSQAASETLERVRDAIDRNDLSAGLGYALADGTVKVELERFAVAIAERFGDRTFLPLAAKDTDG
KTFESVTAGMTPGQKAEVQSAWNSMRTIQQLSAHERTTEALKQAETMRQTQSQGLSQK
MAVPHFSVSVVARGSGRSAVLSAAYRHCAKMEYEREARVIDYTRKQDLLHEEFVVPADTPEWLRSMISAR
SVALRSEAFWNKVEEFEKRADAQLARDVTIALPIELTAGQNIALVRDFVERHVTAKGMVADWVYHDAPGN
PHVHLMTTLRPLTEDGFGSKRIAVTGPDGKPMRNDAGKIVYELWAGSVDDFNAFRDGWFACQNHHLALAG
FDVRIDGRSFEKQAVDLEPTIHLGVGTKAIERKREQSGRKREAFPSRLERIEIQDARRVENTRRIQRRPE
IVLDLITREKSVFDERDVAKVLYRYIDDGRLFQSLMVRILQSPQALRLERERINLATGTRSPAKYTTREM
IRLEAGMASRAIWLSGRASHGVPGTVIEATFARHPGLSREQQTAIEHAAGAGRIAVVIGRAGAGKTTMMK
AGREAWEGAGYRVVGGALAGKATEGLEKEAGIQSRTLSSWELRWKQGRDQLDKKTVFILDEAGMVSSRQM
ALFVEAVTRAGAKLVLVGDPEQLQPIEAGAAFRAIADRTGYAELETIYRQREQWMRDASLDLARGSVGKA
VDAYTAHGRMIGSRLKDEAVESLIAAWDRDHDPSKTSLILAHLRRDVRMLNDMARARLVERGVVADGFSF
TTEDGVRMFAAGDQIVFLRNEGSLGVRNGMLARVLEAVPGRIVAETGEGEFRRRVTVEQRFYNNLDHGYA
TTIHKAQGATVDRVKVLASLSLDRHLTYVAMTRHREDLAVYYGNRSFAKAGGLVPILSRQNAKETTLDYE
NASFYRQALRFAETRGLHLVNVARTIARDRLEWTLRQKQKLVDLGARLAAIGRAPGLLRGSKNQSIPDPI
KEARPMVSGITIFPKSLEQAVEDKLAGDPGLKKQWQDVSSRFHLVYAQPEAAFNAINVDAMIIDQSVAKS
TLARIAGEPESFGALNGKTGLLASRMEKQKRETAIANVPALARNLERYLRERAEAELKHEREERAARLKV
SIDIPALSQAASETLERVRDAIDRNDLSAGLGYALADGTVKVELERFAVAIAERFGDRTFLPLAAKDTDG
KTFESVTAGMTPGQKAEVQSAWNSMRTIQQLSAHERTTEALKQAETMRQTQSQGLSQK
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2766 | GenBank | WP_137006358 |
| Name | traG_CFBP7129_RS27415_pAtCFBP7129a |
UniProt ID | _ |
| Length | 650 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 650 a.a. Molecular weight: 70123.35 Da Isoelectric Point: 9.3553
>WP_137006358.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MTITRVVLVVVPATLMILIAAGMTGIEEWLAGFGKTGAARLALGRAGIALPYVVSAAIGTLLLFATAGSI
NIRQAGWGVVAGSTGTILIAAMHETTRLAALTSRLPADTTVFTYLDPATSIGAGMALICTCFALRVAFIG
NAAFARTEPKRIQGKRSLHGEADWMKLGEAAKLFPEAGGIVIGERYRVDKDNAAGRAFRADSPETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWNGTLIALDPSKEVAPMVFAHRHNAGRDVLVLD
PKVPDIGFNVLDWIGDFGGTKEEDIASVASWIMSDSGGARGARDDFFRASALQLLTAMIADVCLSGHTEK
RNQTLRQVRKNLSVPEQTLREQLQVIYSQSDSNFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPTYA
ALVSGTTFSTRKIAAGDTDIFINIDLKTLETHSGLVRVIIGSLLNAIYNRDGKIKSRALFLLDEVALLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWLSFAAINDPETAEYISRRCGITT
VEIDRISSSSQAKGLSRTRSKQLAARPLIQAHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVS
SNRFQPIGSDRSPPDPSGRD
MTITRVVLVVVPATLMILIAAGMTGIEEWLAGFGKTGAARLALGRAGIALPYVVSAAIGTLLLFATAGSI
NIRQAGWGVVAGSTGTILIAAMHETTRLAALTSRLPADTTVFTYLDPATSIGAGMALICTCFALRVAFIG
NAAFARTEPKRIQGKRSLHGEADWMKLGEAAKLFPEAGGIVIGERYRVDKDNAAGRAFRADSPETWGAGG
KSPLLCFDGSFGSSHGIVFAGSGGFKTTSVTIPTALKWNGTLIALDPSKEVAPMVFAHRHNAGRDVLVLD
PKVPDIGFNVLDWIGDFGGTKEEDIASVASWIMSDSGGARGARDDFFRASALQLLTAMIADVCLSGHTEK
RNQTLRQVRKNLSVPEQTLREQLQVIYSQSDSNFVKENVAAFVNMTPETFSGVYANAVKETHWLSYPTYA
ALVSGTTFSTRKIAAGDTDIFINIDLKTLETHSGLVRVIIGSLLNAIYNRDGKIKSRALFLLDEVALLGY
MRILETARDAGRKYGITLTMIYQSIGQMRETYGGRDAASKWFESASWLSFAAINDPETAEYISRRCGITT
VEIDRISSSSQAKGLSRTRSKQLAARPLIQAHEVLRMRADEQIVFTAGNAPLRCGRAIWFRRDDMKACVS
SNRFQPIGSDRSPPDPSGRD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2767 | GenBank | WP_137006323 |
| Name | t4cp2_CFBP7129_RS27965_pAtCFBP7129a |
UniProt ID | _ |
| Length | 812 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 812 a.a. Molecular weight: 91192.04 Da Isoelectric Point: 5.8051
>WP_137006323.1 conjugal transfer protein TrbE [Agrobacterium tumefaciens]
MVALRRFRKTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSDSATNTERNELSRQINAILSRIG
SGWMIHVEAMRVPTSSYPSPDRCRFPDPVTRAIDEERRAHFAREQGHFESKHALVLTYRPPEGRKSALSK
YIYSDTESRKTPYAETVLLFFRNMTREIEQYLSSIISIARMKTREVAGHDGEKATRYDDLLQFVRFCITG
ENHPVRLPSVPMYLDWVATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDQMPLSYRWSSRFIF
MDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVVETEDAIAQASSQLVSYGYYTPVIIL
FDENREALSEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYANVREPLINTNNLADLIPLNSVWS
GNPAAPCPFYPENSPPLMLVATGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAADGDHYEIGGDESDEDRALSFCPLFDLSSDADRAWAAEWLEMLTALQGVTITPDHRN
AISRQVMLMATAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQCFEIEHLMTLG
ERTLVPVLTYLFRRIEKRLDGSPSLIILDEAWLMLGHPVFRAKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCLTKICLPNGAALEPGVREFYERIGFNDRQIEIVSSAIPKREYYVATPVGRRLFEMSLGP
IALSFVGVSGKEDLKRIRALRRDHGEGWPIKWLETRGIENAI
MVALRRFRKTGPSFADLVPYAGLVDNGILLLKDGSLMAGWYFAGPDSDSATNTERNELSRQINAILSRIG
SGWMIHVEAMRVPTSSYPSPDRCRFPDPVTRAIDEERRAHFAREQGHFESKHALVLTYRPPEGRKSALSK
YIYSDTESRKTPYAETVLLFFRNMTREIEQYLSSIISIARMKTREVAGHDGEKATRYDDLLQFVRFCITG
ENHPVRLPSVPMYLDWVATAELEHGLTPKVENRFLGVVAIDGLPAESWPGILNSLDQMPLSYRWSSRFIF
MDAEEARQKLERTRKKWQQKVRPFFDQLFQTQSRSVDQDAMTMVVETEDAIAQASSQLVSYGYYTPVIIL
FDENREALSEKAEAVRRLIQAEGFGARIETLNATDAFLGSLPGNWYANVREPLINTNNLADLIPLNSVWS
GNPAAPCPFYPENSPPLMLVATGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLALIAAQFRRYENAQIFA
FDKGSSLLPLTLAADGDHYEIGGDESDEDRALSFCPLFDLSSDADRAWAAEWLEMLTALQGVTITPDHRN
AISRQVMLMATAPGRSLSDFVSGVQMREIKDALHHYTVDGPMGQLLDAEEDGLTLGAFQCFEIEHLMTLG
ERTLVPVLTYLFRRIEKRLDGSPSLIILDEAWLMLGHPVFRAKIREWLKVLRKANCAVVLATQSISDAER
SGIIDVLKESCLTKICLPNGAALEPGVREFYERIGFNDRQIEIVSSAIPKREYYVATPVGRRLFEMSLGP
IALSFVGVSGKEDLKRIRALRRDHGEGWPIKWLETRGIENAI
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 463342..472634
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CFBP7129_RS27910 (CFBP7129_27910) | 459380..460453 | - | 1074 | WP_137006316 | class I SAM-dependent methyltransferase | - |
| CFBP7129_RS32310 (CFBP7129_27915) | 460714..461433 | - | 720 | WP_137006317 | LuxR family transcriptional regulator | - |
| CFBP7129_RS27920 (CFBP7129_27920) | 461756..461971 | - | 216 | WP_137006318 | helix-turn-helix transcriptional regulator | - |
| CFBP7129_RS27925 (CFBP7129_27925) | 462166..462468 | + | 303 | WP_233284861 | transcriptional repressor TraM | - |
| CFBP7129_RS27930 (CFBP7129_27930) | 462460..463164 | - | 705 | WP_137006319 | autoinducer binding domain-containing protein | - |
| CFBP7129_RS27935 (CFBP7129_27935) | 463342..464634 | - | 1293 | WP_137006374 | IncP-type conjugal transfer protein TrbI | virB10 |
| CFBP7129_RS27940 (CFBP7129_27940) | 464645..464950 | - | 306 | WP_337721748 | hypothetical protein | - |
| CFBP7129_RS27945 (CFBP7129_27945) | 465081..465893 | - | 813 | WP_137006375 | P-type conjugative transfer protein TrbG | virB9 |
| CFBP7129_RS27950 (CFBP7129_27950) | 465911..466573 | - | 663 | WP_137006321 | conjugal transfer protein TrbF | virB8 |
| CFBP7129_RS27955 (CFBP7129_27955) | 466601..467728 | - | 1128 | WP_233284862 | P-type conjugative transfer protein TrbL | virB6 |
| CFBP7129_RS27960 (CFBP7129_27960) | 467769..468527 | - | 759 | WP_233284886 | P-type conjugative transfer protein TrbJ | virB5 |
| CFBP7129_RS27965 (CFBP7129_27965) | 468562..471000 | - | 2439 | WP_137006323 | conjugal transfer protein TrbE | virb4 |
| CFBP7129_RS27970 (CFBP7129_27970) | 471013..471312 | - | 300 | WP_137006324 | conjugal transfer protein TrbD | virB3 |
| CFBP7129_RS27975 (CFBP7129_27975) | 471305..471667 | - | 363 | WP_137006325 | TrbC/VirB2 family protein | virB2 |
| CFBP7129_RS27980 (CFBP7129_27980) | 471657..472634 | - | 978 | WP_137006326 | P-type conjugative transfer ATPase TrbB | virB11 |
| CFBP7129_RS27985 (CFBP7129_27985) | 472591..473268 | - | 678 | WP_137006327 | acyl-homoserine-lactone synthase TraI | - |
Host bacterium
| ID | 4366 | GenBank | NZ_CP039924 |
| Plasmid name | pAtCFBP7129a | Incompatibility group | - |
| Plasmid size | 473557 bp | Coordinate of oriT [Strand] | 361545..361598 [-] |
| Host baterium | Agrobacterium tumefaciens strain CFBP7129 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | arsC, arsH |
| Degradation gene | bphC |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA9 |