Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103919 |
| Name | oriT_pAtCFBP6623a |
| Organism | Agrobacterium tumefaciens strain CFBP6623 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP039905 (40280..40308 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pAtCFBP6623a
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file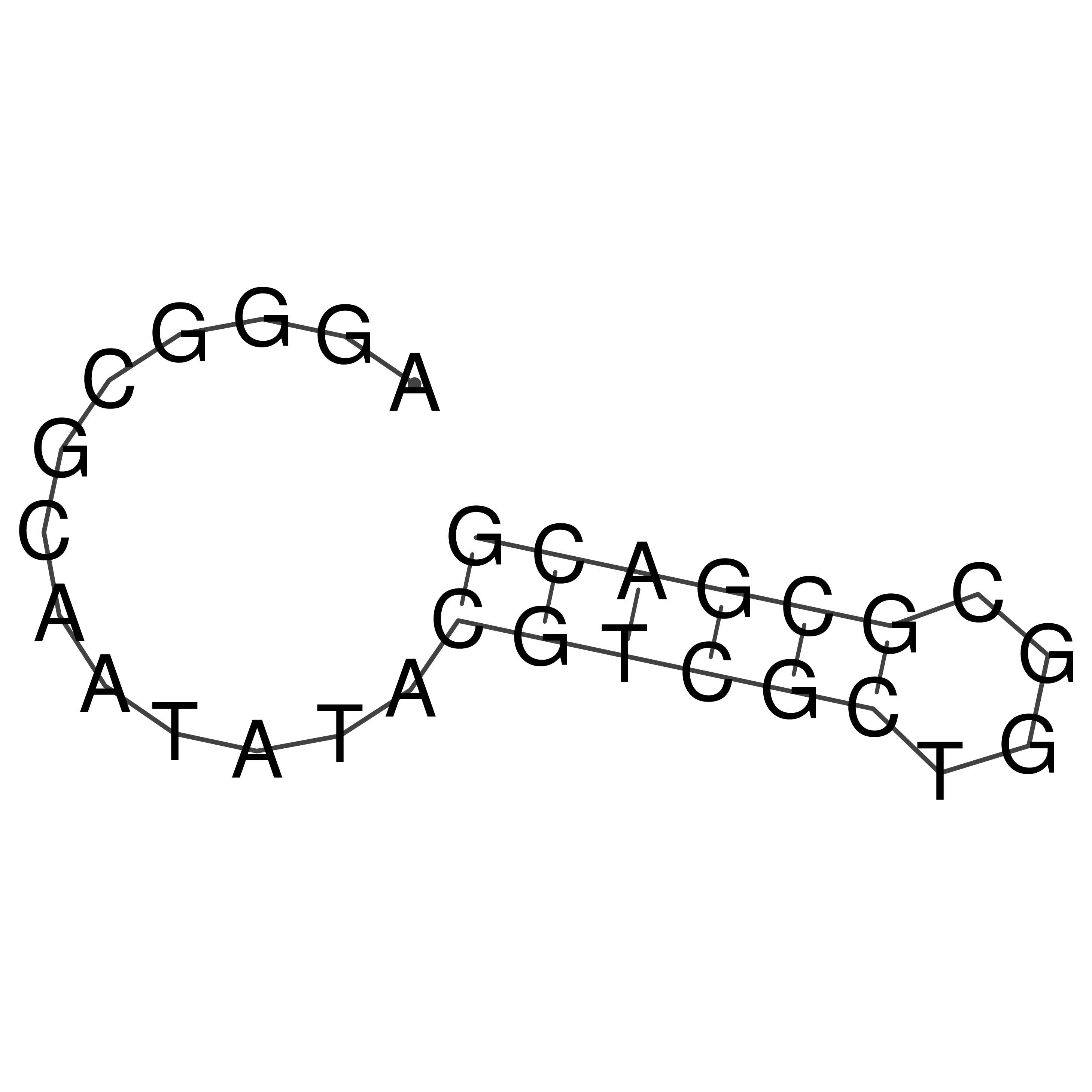
Relaxase
| ID | 2911 | GenBank | WP_062653651 |
| Name | traA_CFBP6623_RS25290_pAtCFBP6623a |
UniProt ID | _ |
| Length | 1538 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1538 a.a. Molecular weight: 170962.07 Da Isoelectric Point: 7.5063
>WP_062653651.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Rhizobium/Agrobacterium group]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQMPAWLQSAVDG
RSVAKASEALWNAVDTFETRADAQLARELIIALPEELTRAENIALVREFVRDNFISKGMIADWVLHDKDG
NPHIHLMTTLRPLTDEGFGPKKVPVLDADGDPLRVVTPDRPNGKIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LSNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDDLVLLKPQQVDTETGKATEPAVFTTREILRIE
FDMAQSAQVSSARRGFVVSDRAIAAAIKKVETQDPEKAFRLDPEQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESNGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWASGRELLGRGDVMVIDEAGMVS
SQQMARVLKIAEEAQAKIVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRDEWARDASRHFARGK
IEESLDAYAQQGRIVETETRADLVERIVADWTAARRELLQKSADGEHPGRLRGDELLVLAHTNVDVQKLN
AFLRKVMMEEGALTGAREFQTARGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVSTGDKR
GDPLLTIHLDNGRDIVISEASYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAASEDFEAKPEWGREPRVDHAAGVTGELVETGEAKFRPHDEDADESPYADVKTDDGTVRLWGVSLPKAL
EDARVSDGDTVTLRKGGVERVKVQVPIFDEKTGQKRFEEREVDRNVWTAKQIETAEVRLERIERESHRPD
VFKQLVERLCRSGAKTTTLDFESEEGYRAHARDFARRRGIDTLAEVAAGMEEGASRRLAWILERKEQVAK
LWERASVALGFAIERERRVSYNEAQNETLAADVGARGKYLIPPTTLFVRSVEEDARRAQLASERWKERDV
ILRPVLEKIYRDPDQALVRLDALASDAGIEPRRLAEDLAVTSDRLGRLRGSDLMVDGRAARDERNIAKAA
MAELLPLARAHATEFRRQAERFGIREQQRRAHMSLSIPALSKQAMARLVEIEAVRDRGGDDAYKTAFAFA
VEDRLLVQEVKAVNDALSGRFGWSAFTAKADATAERSMVERMPEDLAPERREKLTRLFAVIRRFAEEQHL
AERQDRSKIVAGASVALDKETAPMLPMFAAVTEFKTPVEEEARGRALAASHYRHHRAALAETATRIWRDP
AGAIGKIEELVMKSFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERRDALLAVPEAAGRVRSLGA
SYASALEAETRDITEERRRMAVAIPGLSQPAEDALRQLAAEMKKKNAKLAVAVSSLDPRVAQEFTAVSRA
LDERFGRNAILRGEKDVINRVPPAQRRAFEAMHERLKVLQQTVRMQSSQEIIAERQRRVLDRARGVIR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQAGTSFSYRGGASELVHEELALPDQMPAWLQSAVDG
RSVAKASEALWNAVDTFETRADAQLARELIIALPEELTRAENIALVREFVRDNFISKGMIADWVLHDKDG
NPHIHLMTTLRPLTDEGFGPKKVPVLDADGDPLRVVTPDRPNGKIVYKLWAGDKETMKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LSNERSTFDERDIAKALHRTVDDPLDFANIRARLMASDDLVLLKPQQVDTETGKATEPAVFTTREILRIE
FDMAQSAQVSSARRGFVVSDRAIAAAIKKVETQDPEKAFRLDPEQVDAVRHVTGDNGIAAVVGLAGAGKS
TLLAAARVAWESNGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWASGRELLGRGDVMVIDEAGMVS
SQQMARVLKIAEEAQAKIVLVGDAMQLQPIQAGAAFRAITERIGFAELAGVRRQRDEWARDASRHFARGK
IEESLDAYAQQGRIVETETRADLVERIVADWTAARRELLQKSADGEHPGRLRGDELLVLAHTNVDVQKLN
AFLRKVMMEEGALTGAREFQTARGVREFAAGDRIIFLENARFLEPRARRLGPQYVKNGMLGTVVSTGDKR
GDPLLTIHLDNGRDIVISEASYRNIDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAASEDFEAKPEWGREPRVDHAAGVTGELVETGEAKFRPHDEDADESPYADVKTDDGTVRLWGVSLPKAL
EDARVSDGDTVTLRKGGVERVKVQVPIFDEKTGQKRFEEREVDRNVWTAKQIETAEVRLERIERESHRPD
VFKQLVERLCRSGAKTTTLDFESEEGYRAHARDFARRRGIDTLAEVAAGMEEGASRRLAWILERKEQVAK
LWERASVALGFAIERERRVSYNEAQNETLAADVGARGKYLIPPTTLFVRSVEEDARRAQLASERWKERDV
ILRPVLEKIYRDPDQALVRLDALASDAGIEPRRLAEDLAVTSDRLGRLRGSDLMVDGRAARDERNIAKAA
MAELLPLARAHATEFRRQAERFGIREQQRRAHMSLSIPALSKQAMARLVEIEAVRDRGGDDAYKTAFAFA
VEDRLLVQEVKAVNDALSGRFGWSAFTAKADATAERSMVERMPEDLAPERREKLTRLFAVIRRFAEEQHL
AERQDRSKIVAGASVALDKETAPMLPMFAAVTEFKTPVEEEARGRALAASHYRHHRAALAETATRIWRDP
AGAIGKIEELVMKSFAGERIAAAVSNDPAAYGALRGSDRIMDKLLAVGRERRDALLAVPEAAGRVRSLGA
SYASALEAETRDITEERRRMAVAIPGLSQPAEDALRQLAAEMKKKNAKLAVAVSSLDPRVAQEFTAVSRA
LDERFGRNAILRGEKDVINRVPPAQRRAFEAMHERLKVLQQTVRMQSSQEIIAERQRRVLDRARGVIR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2756 | GenBank | WP_062653644 |
| Name | traG_CFBP6623_RS25275_pAtCFBP6623a |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70248.03 Da Isoelectric Point: 9.1224
>WP_062653644.1 MULTISPECIES: Ti-type conjugative transfer system protein TraG [Rhizobium/Agrobacterium group]
MALKGKLHPSLLLTLVPVVVTAITVHIAGWRWTGLATGMSGKTAYWFLRAAPLPALLFGPLAGLLTVWAL
PMHRRRPIAMVSLLCFLGVVGFYALREFGRLAPSVERGLVSWDYALSYLDMVAVVGALGGFMAVAASARI
SSVVPEQVKRARRATFGDADWLSMSAAGKLFPPDGEIVVGERYRVDKEIVHELPFDPNDRSTWGQGGKAP
LLTYGQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTAKLGREVMVLDPT
NPIMGFNVLDGIETSKQKEEDIVGVAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPDYTGRRN
LRSLRQIVSEPEPSVLAMLRDIQENSGSTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GSAFTSSDIVTGTKDVFLNIPASILRSYPGIGRVIIGALINAMIQADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAR
ANRFVKAEP
MALKGKLHPSLLLTLVPVVVTAITVHIAGWRWTGLATGMSGKTAYWFLRAAPLPALLFGPLAGLLTVWAL
PMHRRRPIAMVSLLCFLGVVGFYALREFGRLAPSVERGLVSWDYALSYLDMVAVVGALGGFMAVAASARI
SSVVPEQVKRARRATFGDADWLSMSAAGKLFPPDGEIVVGERYRVDKEIVHELPFDPNDRSTWGQGGKAP
LLTYGQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVVEHRTAKLGREVMVLDPT
NPIMGFNVLDGIETSKQKEEDIVGVAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPDYTGRRN
LRSLRQIVSEPEPSVLAMLRDIQENSGSTFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GSAFTSSDIVTGTKDVFLNIPASILRSYPGIGRVIIGALINAMIQADGGFKRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLMYQSVGQLERHFGKDGATSWIDGCAFASYAAIKALDTARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKEMDQAAR
ANRFVKAEP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 6494..16107
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CFBP6623_RS25065 (CFBP6623_25065) | 2432..3676 | + | 1245 | WP_062653962 | plasmid replication protein RepC | - |
| CFBP6623_RS25070 (CFBP6623_25070) | 3908..4537 | - | 630 | WP_080843285 | RES family NAD+ phosphorylase | - |
| CFBP6623_RS25075 (CFBP6623_25075) | 4534..5319 | - | 786 | WP_062653958 | hypothetical protein | - |
| CFBP6623_RS25080 (CFBP6623_25080) | 5502..5852 | - | 351 | WP_062653986 | MarR family transcriptional regulator | - |
| CFBP6623_RS25085 (CFBP6623_25085) | 5951..6490 | + | 540 | WP_062653956 | hypothetical protein | - |
| CFBP6623_RS25090 (CFBP6623_25090) | 6494..7159 | + | 666 | WP_062653949 | lytic transglycosylase domain-containing protein | virB1 |
| CFBP6623_RS25095 (CFBP6623_25095) | 7156..7455 | + | 300 | WP_062653947 | TrbC/VirB2 family protein | virB2 |
| CFBP6623_RS25100 (CFBP6623_25100) | 7462..7800 | + | 339 | WP_062653945 | type IV secretion system protein VirB3 | virB3 |
| CFBP6623_RS25105 (CFBP6623_25105) | 7793..10159 | + | 2367 | WP_062653942 | VirB4 family type IV secretion system protein | virb4 |
| CFBP6623_RS25110 (CFBP6623_25110) | 10156..10857 | + | 702 | WP_062653941 | P-type DNA transfer protein VirB5 | virB5 |
| CFBP6623_RS25115 (CFBP6623_25115) | 10854..11084 | + | 231 | WP_062653939 | EexN family lipoprotein | - |
| CFBP6623_RS25120 (CFBP6623_25120) | 11088..12020 | + | 933 | WP_062653937 | type IV secretion system protein | virB6 |
| CFBP6623_RS25125 (CFBP6623_25125) | 12060..12341 | + | 282 | WP_062653935 | hypothetical protein | - |
| CFBP6623_RS25130 (CFBP6623_25130) | 12343..13014 | + | 672 | WP_062653931 | virB8 family protein | virB8 |
| CFBP6623_RS25135 (CFBP6623_25135) | 13011..13868 | + | 858 | WP_145980565 | P-type conjugative transfer protein VirB9 | virB9 |
| CFBP6623_RS25140 (CFBP6623_25140) | 13877..15052 | + | 1176 | WP_080843287 | type IV secretion system protein VirB10 | virB10 |
| CFBP6623_RS25145 (CFBP6623_25145) | 15061..16107 | + | 1047 | WP_062653600 | P-type DNA transfer ATPase VirB11 | virB11 |
| CFBP6623_RS25150 (CFBP6623_25150) | 16104..16535 | - | 432 | WP_062653602 | winged helix-turn-helix domain-containing protein | - |
| CFBP6623_RS25155 (CFBP6623_25155) | 17017..19149 | + | 2133 | WP_062653604 | ParB/RepB/Spo0J family partition protein | - |
| CFBP6623_RS25160 (CFBP6623_25160) | 19220..19876 | + | 657 | WP_062653662 | hypothetical protein | - |
| CFBP6623_RS25165 (CFBP6623_25165) | 20022..20444 | + | 423 | WP_062653606 | antirestriction protein | - |
Host bacterium
| ID | 4359 | GenBank | NZ_CP039905 |
| Plasmid name | pAtCFBP6623a | Incompatibility group | - |
| Plasmid size | 260250 bp | Coordinate of oriT [Strand] | 40280..40308 [+] |
| Host baterium | Agrobacterium tumefaciens strain CFBP6623 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | golT |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | - |