Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103892 |
| Name | oriT_pII |
| Organism | Agrobacterium tumefaciens strain 183 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP029047 (64971..65006 [-], 36 nt) |
| oriT length | 36 nt |
| IRs (inverted repeats) | 22..27, 31..36 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 36 nt
>oriT_pII
GGTTCCAAGGGCGCAATTATACGTCGCGATGCGACG
GGTTCCAAGGGCGCAATTATACGTCGCGATGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file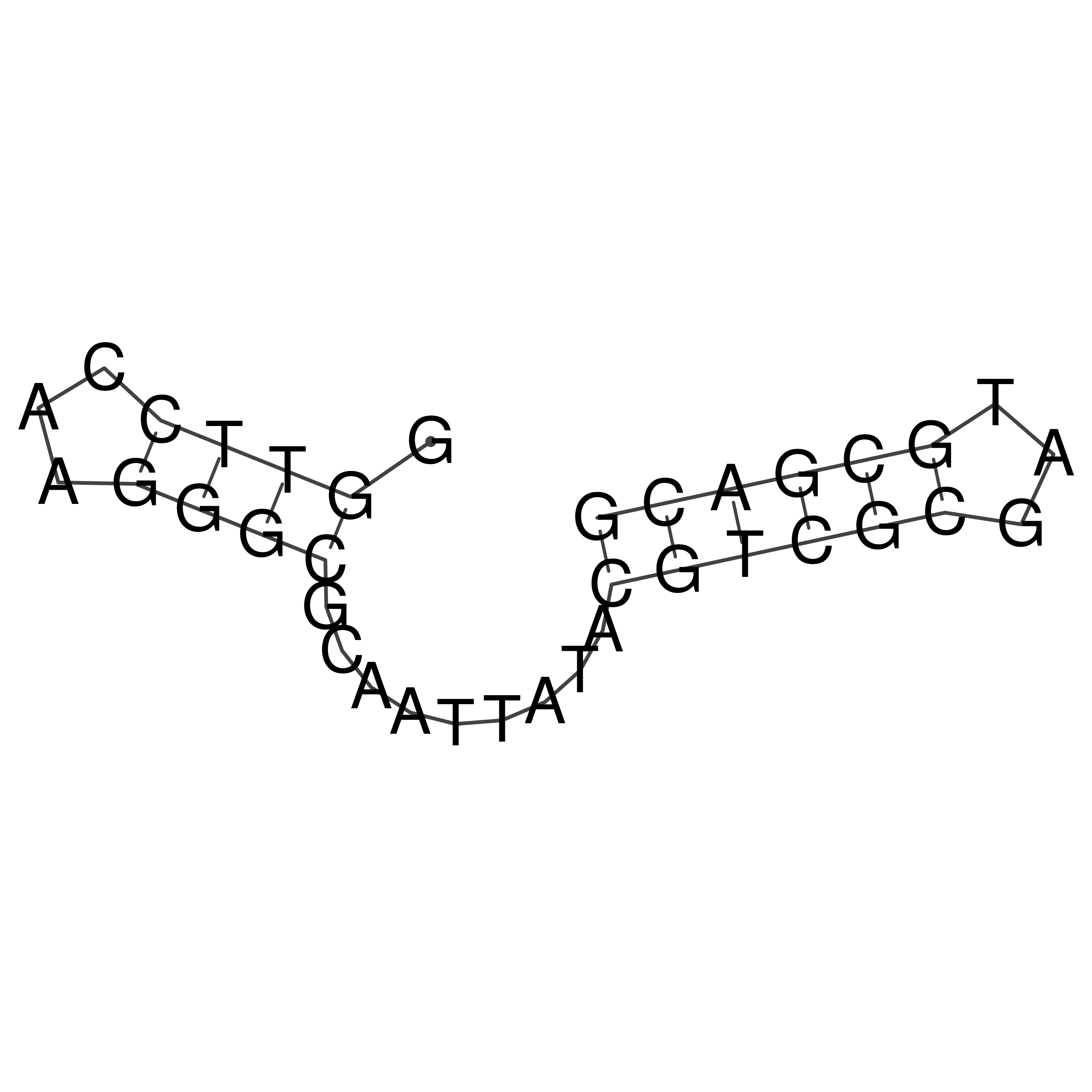
Relaxase
| ID | 2895 | GenBank | WP_128287276 |
| Name | traA_DC439_RS25290_pII |
UniProt ID | _ |
| Length | 1100 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1100 a.a. Molecular weight: 123422.11 Da Isoelectric Point: 9.9780
>WP_128287276.1 MULTISPECIES: Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens complex]
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFLAPADAPKWVRSLIADR
SVSGAVEAFWNKVEAFEKRADAQLARDLTIALPIELSVEQNIVLVRDFVEKHILAKGMFADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGEDGRPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAEQQGVRPELERIALNEERRSENTRRILNNPAIVLDL
ITREKSVFDERDIAKVLSRYVDDPAVFQQLMLRIILNPEVLRLQRDTIDFATGEKVPARYSTRTMIRLEA
TMSNQALWLSGKETHAVSASVLDATFRRHVRLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDNRCVFVMDEAGMVASKQMAGFVD
AVVRSGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALNAYG
ANAKITGERLKDEAVKSLITDWNHSYDPKKTTLILAHLRRDVRMLNVMAREKLVERSIVGDGHVFKTADG
IRQFDTGDQIVFLKNDTSLGVKNGMIGHVVEAAANRIVAVTGEGEHRRHVVVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRKNAKETTLDYERGQLY
REALRFAENRGLRIVQVARTLLRDRLDWTLRQKAKLTDLGQRLAAFAERLGLNQSLKAQTMKEAAPMVAG
IKTFSRSVADTVGDQLGADPALKRQWEEVSARFRYVFADPETAFRAMNFDVVLADRQAAKQVLQKLEAEP
ASIGPLNGKAGILASKAEREARRIAEVNVPALKRDFEQYLRMREMVAQRLQTDEQVLRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAAMAYALSNRETKLEIEGFNQAVTERFGERTLLSNAAREPSGKLFEKLSEG
MKPEQKEQLKQAWPVMRTAQQLAAHERTVHSLKLAEEQGLTQRQTPVLKQ
MAIAHFSASIVSRGSGRSVVLSAAYRHCAKMEYEREARTIDYTRKQGLLHEEFLAPADAPKWVRSLIADR
SVSGAVEAFWNKVEAFEKRADAQLARDLTIALPIELSVEQNIVLVRDFVEKHILAKGMFADWVYHDNPGN
PHIHLMTTLRPLTEDGFGSKKVAVTGEDGRPVRTKSGKILYELWAGSTDDFNVLRDGWFERLNHHLALGG
IDLRIDGRSYEKQGIDLEPTIHLGVGAKAIERKAEQQGVRPELERIALNEERRSENTRRILNNPAIVLDL
ITREKSVFDERDIAKVLSRYVDDPAVFQQLMLRIILNPEVLRLQRDTIDFATGEKVPARYSTRTMIRLEA
TMSNQALWLSGKETHAVSASVLDATFRRHVRLSEEQKTAIERIAGPARIAAVVGRAGAGKTTMMKAAREA
WELAGYRVVGGALAGKAAEGLEKEAGIQSRTLASWELRWNRGRDVLDNRCVFVMDEAGMVASKQMAGFVD
AVVRSGAKIVLVGDPEQLQPIEAGAAFRAIVDRIGYAELETIYRQREDWMRKASLDLARGNVEKALNAYG
ANAKITGERLKDEAVKSLITDWNHSYDPKKTTLILAHLRRDVRMLNVMAREKLVERSIVGDGHVFKTADG
IRQFDTGDQIVFLKNDTSLGVKNGMIGHVVEAAANRIVAVTGEGEHRRHVVVEQRFYSNLDHGYATTIHK
SQGATVDRVKVLASLSLDRHLTYVAMTRHREDLQLYYGTRSFSFNGGLAKVLSRKNAKETTLDYERGQLY
REALRFAENRGLRIVQVARTLLRDRLDWTLRQKAKLTDLGQRLAAFAERLGLNQSLKAQTMKEAAPMVAG
IKTFSRSVADTVGDQLGADPALKRQWEEVSARFRYVFADPETAFRAMNFDVVLADRQAAKQVLQKLEAEP
ASIGPLNGKAGILASKAEREARRIAEVNVPALKRDFEQYLRMREMVAQRLQTDEQVLRQRVSIDIPALSP
AARVVLERVRDAIDRNDLPAAMAYALSNRETKLEIEGFNQAVTERFGERTLLSNAAREPSGKLFEKLSEG
MKPEQKEQLKQAWPVMRTAQQLAAHERTVHSLKLAEEQGLTQRQTPVLKQ
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2739 | GenBank | WP_128287328 |
| Name | t4cp2_DC439_RS25595_pII |
UniProt ID | _ |
| Length | 820 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 820 a.a. Molecular weight: 91545.49 Da Isoelectric Point: 5.9678
>WP_128287328.1 conjugal transfer protein TrbE [Agrobacterium tumefaciens]
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRMPTADYPKESDCHFPDLVTRAIDAERRAHFQKERGHFESRHALILTWRPPEPRRSGLTR
YIYSDMASRSATYADKALETFLTSIREVEQYLANVVSIRRMVTRETSELGGFRIARYDELFQFIRFCVTG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLIENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDAEEARTKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVL
FEEDQARLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLVPLNSVWS
GSPVAPCPFYPTGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYSGAQIFA
FDKGGSMLPLTLGIDGDHYQIGGDATGGGEGKALSFCPLADLTTDGDRAFASEWIEMLVALQGVTITPDY
RNAISRQVGLMAKSRGRSLSDFVSGVQMREIKDALHHYTVDGQMGQLLDAEEDGLALGSFQCFEIEELMN
MGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDA
ERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKRDYYVVSPEGRRLFDMAL
GPVALSFVGASGKEHLKQIRSLHSEHGANWPLHWLQQRGIAHADTLFPAD
MVALKSFRHSGPSFADLVPYAGLVDNGVILLKDGSLMAGWYFAGPDSESSTDAERNEVSRQINAILSRLG
SGWMIQVEAVRMPTADYPKESDCHFPDLVTRAIDAERRAHFQKERGHFESRHALILTWRPPEPRRSGLTR
YIYSDMASRSATYADKALETFLTSIREVEQYLANVVSIRRMVTRETSELGGFRIARYDELFQFIRFCVTG
ENHPVRLPEIPMYLDWLVTAELQHGLTPLIENRFLGVVAIDGLPAESWPGILNALDLMPLTYRWSSRFVF
LDAEEARTKLERTRKKWQQKVRPFFDQLFQTQSRSLDQDAMLMVAETEDAIAEASSQLVAYGYYTPVIVL
FEEDQARLQEKCEAIRRLIQAEGFGARIETLNATDAFLGSLPGVSYANIREPLINTRNLADLVPLNSVWS
GSPVAPCPFYPTGSPPLMQVASGSTPFRLNLHVDDVGHTLIFGPTGSGKSTLLSLIAAQFRRYSGAQIFA
FDKGGSMLPLTLGIDGDHYQIGGDATGGGEGKALSFCPLADLTTDGDRAFASEWIEMLVALQGVTITPDY
RNAISRQVGLMAKSRGRSLSDFVSGVQMREIKDALHHYTVDGQMGQLLDAEEDGLALGSFQCFEIEELMN
MGERNLVPVLTYLFRRIEKRLTGAPSLIILDEAWLMLGHPVFRDKIREWLKVLRKANCAVVLATQSISDA
ERSGIIDVLKESCPTKICLPNGAAREPGTREFYERIGFNERQIEIVATAIPKRDYYVVSPEGRRLFDMAL
GPVALSFVGASGKEHLKQIRSLHSEHGANWPLHWLQQRGIAHADTLFPAD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 4332 | GenBank | NZ_CP029047 |
| Plasmid name | pII | Incompatibility group | - |
| Plasmid size | 140967 bp | Coordinate of oriT [Strand] | 64971..65006 [-] |
| Host baterium | Agrobacterium tumefaciens strain 183 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | - |
| Metal resistance gene | - |
| Degradation gene | benA, benB, benC |
| Symbiosis gene | - |
| Anti-CRISPR | - |