Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103891 |
| Name | oriT_pI |
| Organism | Agrobacterium tumefaciens strain 183 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP029046 (145816..145844 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) 4..9, 20..25 (GCGCAA..TTGCGC) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_pI
AGGGCGCAATATACGTCGCTTGCGCGACG
AGGGCGCAATATACGTCGCTTGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file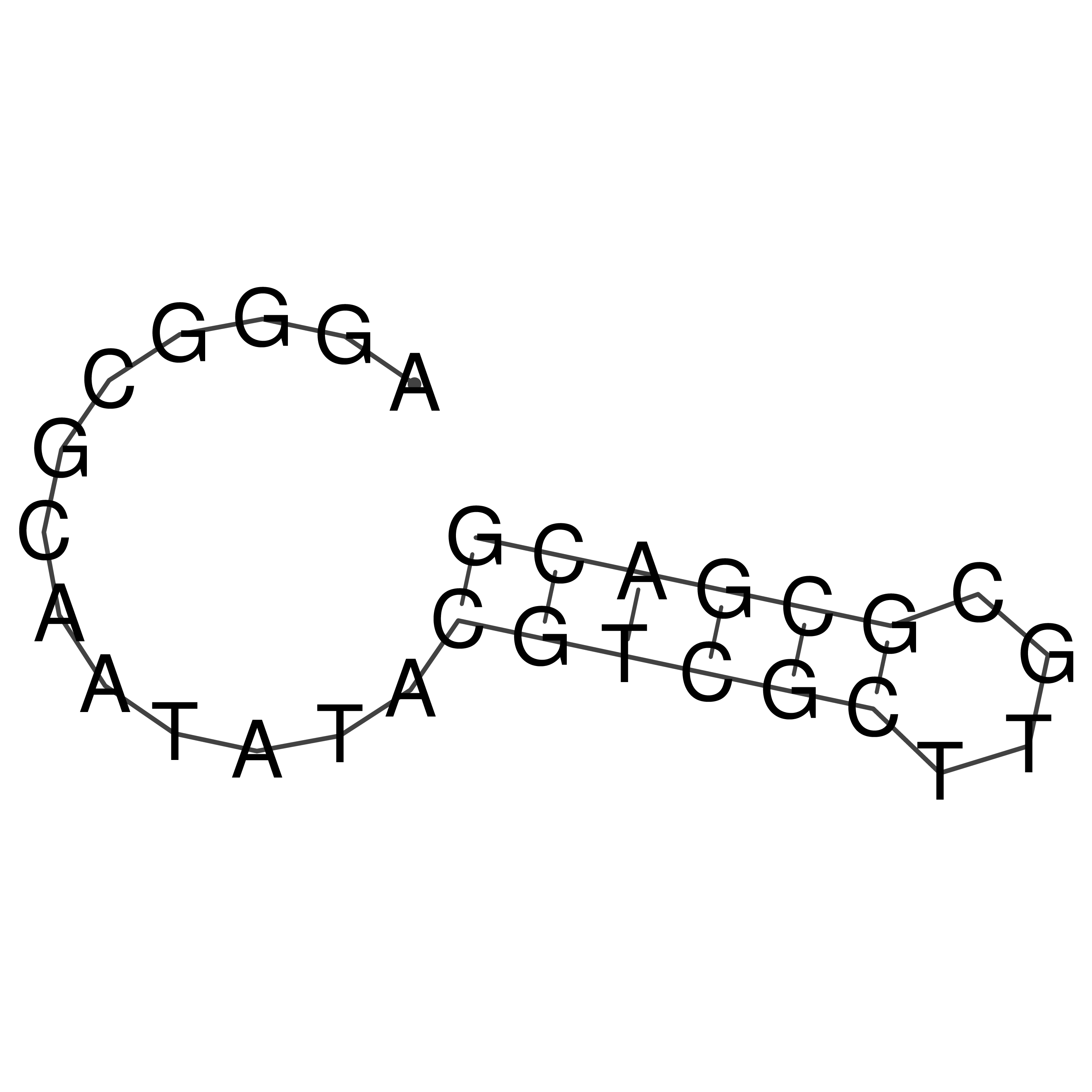
Relaxase
| ID | 2894 | GenBank | WP_128287202 |
| Name | traA_DC439_RS24680_pI |
UniProt ID | _ |
| Length | 1545 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1545 a.a. Molecular weight: 170737.35 Da Isoelectric Point: 7.7226
>WP_128287202.1 Ti-type conjugative transfer relaxase TraA [Agrobacterium tumefaciens]
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQIGSSFSYRGGARELVHEELALPDKVPTWLKTAVDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENISLVREFVADSFTSKGMVADWVFHDKDG
NPHIHLMMTLRPLTEEGFGPKKVQVAGADGEPLRVITPDRPNGKIVYKIWAGDKETMKGWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LANERSTFDERDIAKALHRYVDDPADFANIRARLMTSNDLVLLKPQRMDDQSGKVTEPAVFTTREILRIE
FDMAQSAEVLSKRTGFGVADAKIAAAVRQVETADPEKPFKLDAEQVDAVRHVTGENAVAAVVGLAGAGKS
TLLAAARVAWERDHRGVFGAALAGKAAEGLEDSSGIKSRTLASWELAWANGCDLLQRGNVFVIDEAGMVS
SQQMARVLKVVEEAGAKAVLVGDAMQLQPIQAGAAFRAIGDRIGFAELAGVRRQREEWAREASRLFARGE
VEKGLDAYAAHGRLVESETRDEIIGRMVSDWADARRALVQASRDQENPPRLRGDELLVLAHTNDDVRKLN
EALRKVMLDDGALTGAREFHTARGAREFATGDRIIFLENARFLEPRAKRLGPQYVKNGMLGTIVATGDKR
AGPLLSVRLDNGRDVIISEDSYTNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAREDFQPKLEWGRAARADHAAGVTGELVETGLAKFRPNDEDADESPYADMKTDDGAVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTVTVPVIDENTGKKSFEERTVDRNVWTAKLKETADVRRERIERESHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYQAHARDFARRRGIDMAAGVAAGIEEGLSRRFTWIAEKREQVA
RLWERASVALGFAIERERSVSYNEARAATVSADIPVGGYHLIPPTTTFTRSVEHDARRAQLLSERWKERE
AILRPVLDKVYRDPDAALIRLNALASNSNIEPRKLADDLAMAPNRLGRLRGSDLMVDGRAARDERSAAMA
ALNDLVPLARAHATEFRRNEDRFQDREKTRRAHMALSIPAISKRAMARLVEIEAVRKQGGDDAYKTAFSF
AAEDREIVQEVKAVSDALNARFGWSAFTAKADAAAERNVAERMPEDLPDDRREKLVRLFEVVRHFSDEQH
LAEKQDRSKIVAGASVEAGKESMAMLPMLAAVTEFKTPVDEEARTRALAVPHYRHHLAALAETAGRIWRD
PAGAVGKIEELIVKGIASERIAAAVTNDPAAYGALRGSDRIMEKLLAVGRERKGALQAVPEAASRVRSLG
ASWASALDAESEAIREERRRMSVAIPGVSPAAEGALLHLVADLKKQRQEKAKSARIETAARSLDPDIARE
FAAVSRALDERFGRNAILRGETDVINRVSPAQRRAFEAMRERLQVVQQTVRTHGSQEIISERQRRAIDRA
RGLTR
MAIMFVRAQVISRGAGRSIVSAAAYRHRARMMDEQIGSSFSYRGGARELVHEELALPDKVPTWLKTAVDG
KTVAAASEALWNAVDAFETRADAQLARELIIALPEELTRAENISLVREFVADSFTSKGMVADWVFHDKDG
NPHIHLMMTLRPLTEEGFGPKKVQVAGADGEPLRVITPDRPNGKIVYKIWAGDKETMKGWKIAWAETANR
HLALAGHDIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGVEMYFAPADLARRQEMADRLLAEPELLLKQ
LANERSTFDERDIAKALHRYVDDPADFANIRARLMTSNDLVLLKPQRMDDQSGKVTEPAVFTTREILRIE
FDMAQSAEVLSKRTGFGVADAKIAAAVRQVETADPEKPFKLDAEQVDAVRHVTGENAVAAVVGLAGAGKS
TLLAAARVAWERDHRGVFGAALAGKAAEGLEDSSGIKSRTLASWELAWANGCDLLQRGNVFVIDEAGMVS
SQQMARVLKVVEEAGAKAVLVGDAMQLQPIQAGAAFRAIGDRIGFAELAGVRRQREEWAREASRLFARGE
VEKGLDAYAAHGRLVESETRDEIIGRMVSDWADARRALVQASRDQENPPRLRGDELLVLAHTNDDVRKLN
EALRKVMLDDGALTGAREFHTARGAREFATGDRIIFLENARFLEPRAKRLGPQYVKNGMLGTIVATGDKR
AGPLLSVRLDNGRDVIISEDSYTNVDHGYAATIHKSQGATVDRTFVLATGMMDQHLTYVSMTRHRDRADL
YAAREDFQPKLEWGRAARADHAAGVTGELVETGLAKFRPNDEDADESPYADMKTDDGAVHRLWGVSLPKA
LEEGGVSEGDTVTLRKDGVERVTVTVPVIDENTGKKSFEERTVDRNVWTAKLKETADVRRERIERESHRP
ELFKQLVERLGRSGAKTTTLDFESEEGYQAHARDFARRRGIDMAAGVAAGIEEGLSRRFTWIAEKREQVA
RLWERASVALGFAIERERSVSYNEARAATVSADIPVGGYHLIPPTTTFTRSVEHDARRAQLLSERWKERE
AILRPVLDKVYRDPDAALIRLNALASNSNIEPRKLADDLAMAPNRLGRLRGSDLMVDGRAARDERSAAMA
ALNDLVPLARAHATEFRRNEDRFQDREKTRRAHMALSIPAISKRAMARLVEIEAVRKQGGDDAYKTAFSF
AAEDREIVQEVKAVSDALNARFGWSAFTAKADAAAERNVAERMPEDLPDDRREKLVRLFEVVRHFSDEQH
LAEKQDRSKIVAGASVEAGKESMAMLPMLAAVTEFKTPVDEEARTRALAVPHYRHHLAALAETAGRIWRD
PAGAVGKIEELIVKGIASERIAAAVTNDPAAYGALRGSDRIMEKLLAVGRERKGALQAVPEAASRVRSLG
ASWASALDAESEAIREERRRMSVAIPGVSPAAEGALLHLVADLKKQRQEKAKSARIETAARSLDPDIARE
FAAVSRALDERFGRNAILRGETDVINRVSPAQRRAFEAMRERLQVVQQTVRTHGSQEIISERQRRAIDRA
RGLTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2738 | GenBank | WP_128287199 |
| Name | traG_DC439_RS24665_pI |
UniProt ID | _ |
| Length | 637 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 637 a.a. Molecular weight: 70523.40 Da Isoelectric Point: 9.0190
>WP_128287199.1 Ti-type conjugative transfer system protein TraG [Agrobacterium tumefaciens]
MTIRAKPHPSLLLILVPITVTAFAVYVVGWRWPELAAEMTGKAQYWFLRASPVPALLFGPLAGLLTVLAL
PLHRRRPIAMATLICFLSVACLYGLREFGRLSPSVESGAVTWNQALSFIDMVAVMGSVAGFMATAVSARI
STVVSEPVKRARRGTFGDADWLPMSAAGRLFPDDGEIVVGERYRVDKEIVHEMPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSEQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYEGRRN
LRSLRQIVSEPETSVLAMLRDVQQHSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALAC
GNTFKPSDLTRGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESLNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKTMNEAAK
VNRFVKP
MTIRAKPHPSLLLILVPITVTAFAVYVVGWRWPELAAEMTGKAQYWFLRASPVPALLFGPLAGLLTVLAL
PLHRRRPIAMATLICFLSVACLYGLREFGRLSPSVESGAVTWNQALSFIDMVAVMGSVAGFMATAVSARI
STVVSEPVKRARRGTFGDADWLPMSAAGRLFPDDGEIVVGERYRVDKEIVHEMPFDPNDPATWGQGGKAP
LLTYKQDFDSTHMLFFAGSGGYKTTSNVIPTALRYSGPLICLDPSTEVAPMVVEHRRERLGRQVMVLDPT
NPIMGFNVLDGIETSEQKEQDIVGIAHMLLSESVRFESSTGSYFQSQAHNLLTGLLAHVMLSPEYEGRRN
LRSLRQIVSEPETSVLAMLRDVQQHSASKFISETLGVFVNMTEQTFSGVYSTASKDTQWLSLDNYAALAC
GNTFKPSDLTRGKKDVFLNIPASILRSYPGIGRVIVGSLINAMIEADGAFQRRALFMLDEVDLLGYMRVL
EEARDRGRKYGISMMLLYQSVGQLERHFGKDGAVSWIDGCAFASYAAIKALETARNVSAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESLNFQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKTMNEAAK
VNRFVKP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 73484..83077
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| DC439_RS24305 (DC439_24350) | 69155..70027 | - | 873 | WP_128287142 | sugar ABC transporter permease | - |
| DC439_RS24310 (DC439_24355) | 70204..71517 | - | 1314 | WP_128287143 | sugar ABC transporter substrate-binding protein | - |
| DC439_RS24315 (DC439_24360) | 72057..73190 | + | 1134 | WP_233282957 | hypothetical protein | - |
| DC439_RS24320 (DC439_24365) | 73484..74485 | - | 1002 | WP_128287144 | P-type DNA transfer ATPase VirB11 | virB11 |
| DC439_RS24325 (DC439_24370) | 74494..75666 | - | 1173 | WP_128287145 | type IV secretion system protein VirB10 | virB10 |
| DC439_RS24330 (DC439_24375) | 75680..76534 | - | 855 | WP_161974293 | P-type conjugative transfer protein VirB9 | virB9 |
| DC439_RS24335 (DC439_24380) | 76531..77202 | - | 672 | WP_128287146 | virB8 family protein | virB8 |
| DC439_RS24340 (DC439_24385) | 77206..77472 | - | 267 | WP_128287147 | hypothetical protein | - |
| DC439_RS24345 (DC439_24390) | 77509..78444 | - | 936 | WP_128287148 | type IV secretion system protein | virB6 |
| DC439_RS24350 (DC439_24395) | 78449..78682 | - | 234 | WP_128287149 | EexN family lipoprotein | - |
| DC439_RS24355 (DC439_24400) | 78679..79377 | - | 699 | WP_128287150 | P-type DNA transfer protein VirB5 | virB5 |
| DC439_RS24360 (DC439_24405) | 79377..81758 | - | 2382 | WP_128287151 | VirB4 family type IV secretion system protein | virb4 |
| DC439_RS24365 (DC439_24410) | 81751..82092 | - | 342 | WP_128287152 | type IV secretion system protein VirB3 | virB3 |
| DC439_RS24370 (DC439_24415) | 82095..82394 | - | 300 | WP_128287153 | TrbC/VirB2 family protein | virB2 |
| DC439_RS24375 (DC439_24420) | 82391..83077 | - | 687 | WP_128287266 | lytic transglycosylase domain-containing protein | virB1 |
| DC439_RS24380 (DC439_24425) | 83080..83619 | - | 540 | WP_128287154 | hypothetical protein | - |
| DC439_RS24385 (DC439_24430) | 83713..84075 | + | 363 | WP_128287155 | transcriptional regulator | - |
| DC439_RS27295 (DC439_24435) | 84179..84553 | - | 375 | WP_337587109 | acyl-homoserine-lactone synthase | - |
| DC439_RS24395 (DC439_24440) | 84983..86206 | + | 1224 | WP_170212275 | plasmid partitioning protein RepA | - |
| DC439_RS24400 (DC439_24445) | 86285..87280 | + | 996 | WP_128287156 | plasmid partitioning protein RepB | - |
Host bacterium
| ID | 4331 | GenBank | NZ_CP029046 |
| Plasmid name | pI | Incompatibility group | - |
| Plasmid size | 210169 bp | Coordinate of oriT [Strand] | 145816..145844 [+] |
| Host baterium | Agrobacterium tumefaciens strain 183 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | katA |
| Metal resistance gene | - |
| Degradation gene | benC, benB, benA |
| Symbiosis gene | - |
| Anti-CRISPR | - |