Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103852 |
| Name | oriT_HM006|psymA |
| Organism | Sinorhizobium meliloti strain HM006 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP021830 (402747..402775 [-], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_HM006|psymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file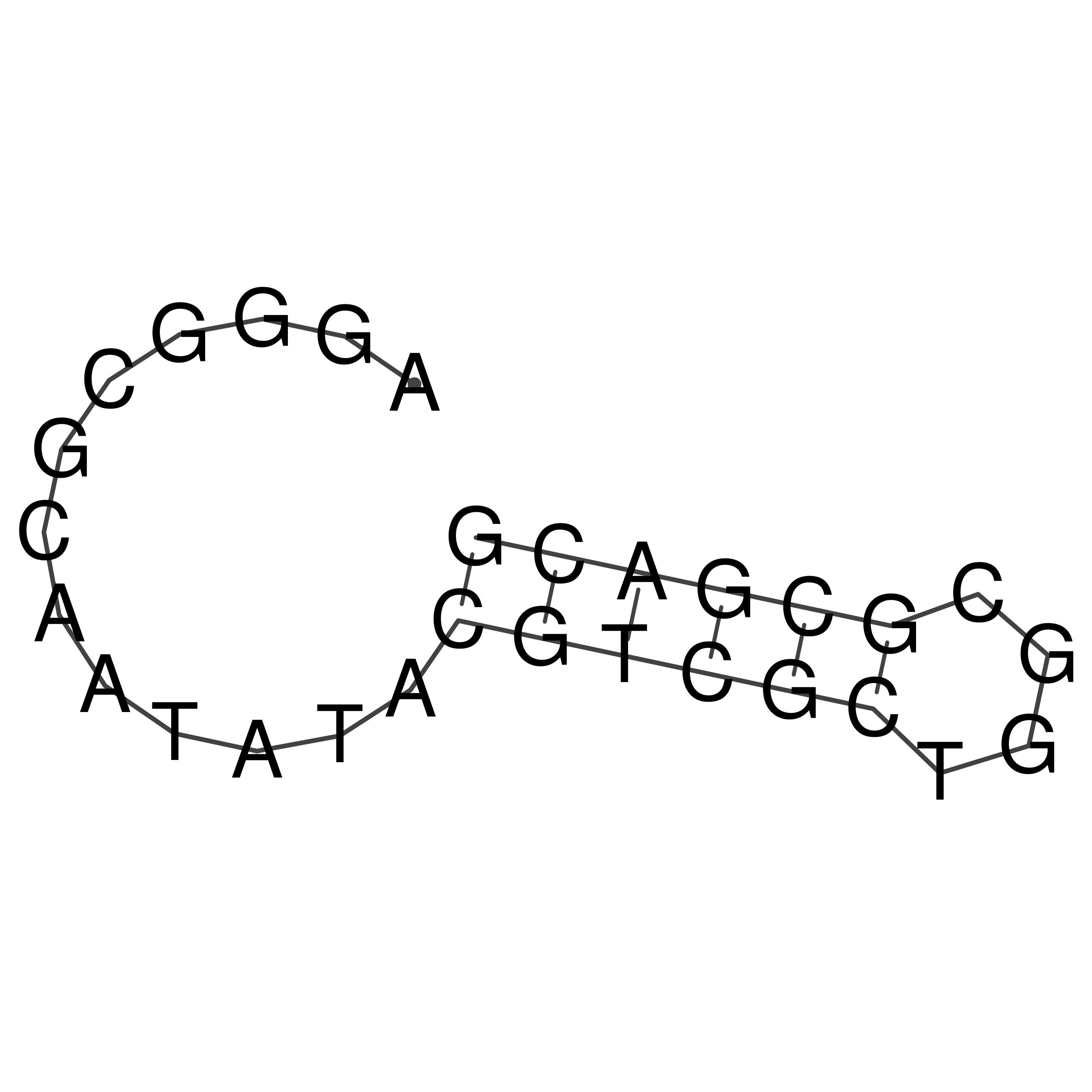
Relaxase
| ID | 2863 | GenBank | WP_088202563 |
| Name | traA_CDO22_RS20195_HM006|psymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 170146.87 Da Isoelectric Point: 9.4212
>WP_088202563.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMTSDELVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMTLSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKDALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMPVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIARALHRYVDDPVDFANIRARLMTSDELVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWEMAWESGREQLQRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARGK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVQRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAGIAEKREQVA
KLWERASVALGFAIERERRVSYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AILHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPDRLGRLRGSELVVDGRAARDERTAATV
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMTLSVPALSKTAMARLVEIEAVRKQGGDDAYRTAFTY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNIAERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKGTFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAAERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKDALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMPVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPHIAREFAKVSR
ALDERFGRNAILRGETDVINRVSPAQRRAFEAMRDRLTILQQAVRIQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2698 | GenBank | WP_027991006 |
| Name | traG_CDO22_RS20210_HM006|psymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70399.34 Da Isoelectric Point: 9.2422
>WP_027991006.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLLILLPVAVTAFAIYVVGWRWPGLAAGMSGNTAYWFLRASPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGLLSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPDDPATWGKGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSSGAYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSQSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSTDIVTGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLLILLPVAVTAFAIYVVGWRWPGLAAGMSGNTAYWFLRASPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGLLSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMSAAGKLFPPDGEIVVGERYRVDKDIVHELPFDPDDPATWGKGGKAP
LLTYSQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVLEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSSGAYFQNQAHNLLTGLLAHVMLSPEYAGRRN
LRSLRQIVSEPEPSVLAMLRDIQEHSQSAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFRSTDIVTGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSENVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
| ID | 2699 | GenBank | WP_088202258 |
| Name | virD4_CDO22_RS20875_HM006|psymA |
UniProt ID | _ |
| Length | 563 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 563 a.a. Molecular weight: 63019.32 Da Isoelectric Point: 9.1751
>WP_088202258.1 type IV secretion system ATPase VirD4 [Sinorhizobium meliloti]
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILLFQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDILAMPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPTPLMIADERF
SHV
MISSKTRPLSIAGSIMCSLAVGFCGASAYSTFRFGFDGRALMTFDILAFWYETPFYLGYTTLFFYRGLAV
VVLTSGAILLFQQMVSVRDRQHHGTARWARVDEMRRAGYLQRYSRISGPVFGKTSGPFWSDYYLTNSEQP
HSLIVAPTRAGKGVGIVIPTLLTYEGSVIALDVKGELFDLTSRARKARGDSVFKLAPLDPERRTNCYNPL
LDILAMPSERQFTEARRLAANLIATKGQSAEGFINGARDLFVAGILACIERGTPTIGAVYDLFAQPGEKY
SLFARLAQETQNKEAQRIFDEMASNDTKILTSYTSVLGDGGLNLWADPLIKAATSRSDFSIYDLRRRRTC
IYLCVSPNDLEVVAPLMRLLFQQVVSILQRSLPGKDEKHEVLFLLDEFKHLGKLEAIETAITTIAGYKGR
FMFIIQSLSALTGTYDESGKQNFLSNTGVQVFMATADDETPVYISKAIGEYTFQARSTSYTQSLTFDRNI
QNSDQGAPLLRPEQVRLLPDKYQIVLIKGQPPLQLRKVRYYSDRALKRIFDSQTGRLPEPTPLMIADERF
SHV
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 273909..283487
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CDO22_RS19505 (CDO22_19510) | 270796..271644 | + | 849 | WP_017264156 | alpha/beta fold hydrolase | - |
| CDO22_RS19510 (CDO22_19515) | 271694..272341 | + | 648 | WP_017264157 | dihydrofolate reductase family protein | - |
| CDO22_RS19515 (CDO22_19520) | 272460..272663 | + | 204 | WP_017264158 | hypothetical protein | - |
| CDO22_RS19520 (CDO22_19525) | 272922..273290 | - | 369 | WP_017264159 | transcriptional regulator | - |
| CDO22_RS36315 (CDO22_19530) | 273387..273905 | + | 519 | WP_017264160 | hypothetical protein | - |
| CDO22_RS19530 (CDO22_19535) | 273909..274580 | + | 672 | WP_017264161 | lytic transglycosylase domain-containing protein | virB1 |
| CDO22_RS19535 (CDO22_19540) | 274577..274876 | + | 300 | WP_003526741 | TrbC/VirB2 family protein | virB2 |
| CDO22_RS19540 (CDO22_19545) | 274881..275222 | + | 342 | WP_017264162 | type IV secretion system protein VirB3 | virB3 |
| CDO22_RS19545 (CDO22_19550) | 275197..277593 | + | 2397 | WP_017264163 | VirB4 family type IV secretion system protein | virb4 |
| CDO22_RS19550 (CDO22_19555) | 277593..278294 | + | 702 | WP_017264164 | P-type DNA transfer protein VirB5 | virB5 |
| CDO22_RS19555 (CDO22_19560) | 278291..278506 | + | 216 | WP_014531545 | EexN family lipoprotein | - |
| CDO22_RS19560 (CDO22_19565) | 278529..279464 | + | 936 | WP_014531546 | type IV secretion system protein | virB6 |
| CDO22_RS19565 (CDO22_19570) | 279461..279775 | + | 315 | WP_017264165 | hypothetical protein | - |
| CDO22_RS19570 (CDO22_19575) | 279780..280451 | + | 672 | WP_010967689 | virB8 family protein | virB8 |
| CDO22_RS19575 (CDO22_19580) | 280451..281305 | + | 855 | WP_014528658 | P-type conjugative transfer protein VirB9 | virB9 |
| CDO22_RS19580 (CDO22_19585) | 281315..282487 | + | 1173 | WP_014990096 | type IV secretion system protein VirB10 | virB10 |
| CDO22_RS19585 (CDO22_19590) | 282498..283487 | + | 990 | WP_010967686 | P-type DNA transfer ATPase VirB11 | virB11 |
| CDO22_RS19590 (CDO22_19595) | 283799..285025 | - | 1227 | WP_010967685 | MFS transporter | - |
| CDO22_RS19595 (CDO22_19600) | 285060..285671 | - | 612 | WP_017264166 | TetR/AcrR family transcriptional regulator | - |
| CDO22_RS19600 (CDO22_19605) | 285923..286600 | + | 678 | WP_017264167 | YoaK family protein | - |
| CDO22_RS19605 (CDO22_19610) | 286848..287870 | + | 1023 | WP_010967682 | alcohol dehydrogenase AdhP | - |
Region 2: 527760..537173
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| CDO22_RS20880 (CDO22_20895) | 523003..523728 | - | 726 | WP_014528527 | response regulator | - |
| CDO22_RS36360 (CDO22_20910) | 524747..525052 | - | 306 | WP_011971014 | hypothetical protein | - |
| CDO22_RS20900 (CDO22_20915) | 525052..526617 | - | 1566 | WP_011971015 | IS66 family transposase | - |
| CDO22_RS20905 (CDO22_20920) | 526689..527036 | - | 348 | WP_011971016 | IS66 family insertion sequence element accessory protein TnpB | - |
| CDO22_RS20910 (CDO22_20925) | 527033..527416 | - | 384 | WP_011971017 | transposase | - |
| CDO22_RS20915 (CDO22_20930) | 527760..528800 | - | 1041 | WP_014528525 | P-type DNA transfer ATPase VirB11 | virB11 |
| CDO22_RS20920 (CDO22_20935) | 528840..529958 | - | 1119 | WP_014528524 | type IV secretion system protein VirB10 | virB10 |
| CDO22_RS20925 (CDO22_20940) | 529955..530836 | - | 882 | WP_014528523 | TrbG/VirB9 family P-type conjugative transfer protein | virB9 |
| CDO22_RS20930 (CDO22_20945) | 530833..531546 | - | 714 | WP_014528522 | type IV secretion system protein VirB8 | virB8 |
| CDO22_RS20935 (CDO22_20950) | 531533..531700 | - | 168 | WP_011970875 | type IV secretion system lipoprotein VirB7 | - |
| CDO22_RS20940 (CDO22_20955) | 531733..532620 | - | 888 | WP_014528521 | type IV secretion system protein | virB6 |
| CDO22_RS20945 (CDO22_20960) | 532701..533384 | - | 684 | WP_014528520 | pilin minor subunit VirB5 | traI |
| CDO22_RS20950 (CDO22_20965) | 533398..535767 | - | 2370 | WP_014528519 | VirB4 family type IV secretion/conjugal transfer ATPase | virb4 |
| CDO22_RS20955 (CDO22_20970) | 535767..536093 | - | 327 | WP_003537077 | type IV secretion system protein VirB3 | virB3 |
| CDO22_RS20960 (CDO22_20975) | 536090..536452 | - | 363 | WP_014528518 | pilin major subunit VirB2 | virB2 |
| CDO22_RS20965 (CDO22_20980) | 536466..537173 | - | 708 | WP_014528517 | type IV secretion system lytic transglycosylase VirB1 | virB1 |
| CDO22_RS35890 | 537382..537540 | + | 159 | WP_170849885 | hypothetical protein | - |
| CDO22_RS20975 (CDO22_20990) | 537692..537883 | - | 192 | Protein_515 | Lrp/AsnC ligand binding domain-containing protein | - |
| CDO22_RS20980 (CDO22_20995) | 537867..538442 | + | 576 | WP_153428644 | type IV secretory system conjugative DNA transfer family protein | - |
| CDO22_RS20985 (CDO22_21000) | 538526..539637 | + | 1112 | Protein_517 | IS256-like element ISRm5 family transposase | - |
| CDO22_RS20990 (CDO22_21005) | 539657..539925 | + | 269 | Protein_518 | hypothetical protein | - |
| CDO22_RS20995 (CDO22_21010) | 540255..541708 | - | 1454 | Protein_519 | PLP-dependent aminotransferase family protein | - |
Host bacterium
| ID | 4292 | GenBank | NZ_CP021830 |
| Plasmid name | HM006|psymA | Incompatibility group | - |
| Plasmid size | 1484605 bp | Coordinate of oriT [Strand] | 402747..402775 [-] |
| Host baterium | Sinorhizobium meliloti strain HM006 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | - |
| Degradation gene | - |
| Symbiosis gene | fixN, fixO, fixG, fixS, nodM, nifN, nodD, nodA, nodB, nodC, nodI, nodJ, nodE, nodF, nodH, nifX, nifE, nifK, nifD, nifH, fixA, fixB, fixC, fixX, nifB, fixU, nifT |
| Anti-CRISPR | - |