Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103849 |
| Name | oriT_B401|pSymB |
| Organism | Sinorhizobium meliloti strain B401 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019484 (867761..867789 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_B401|pSymB
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file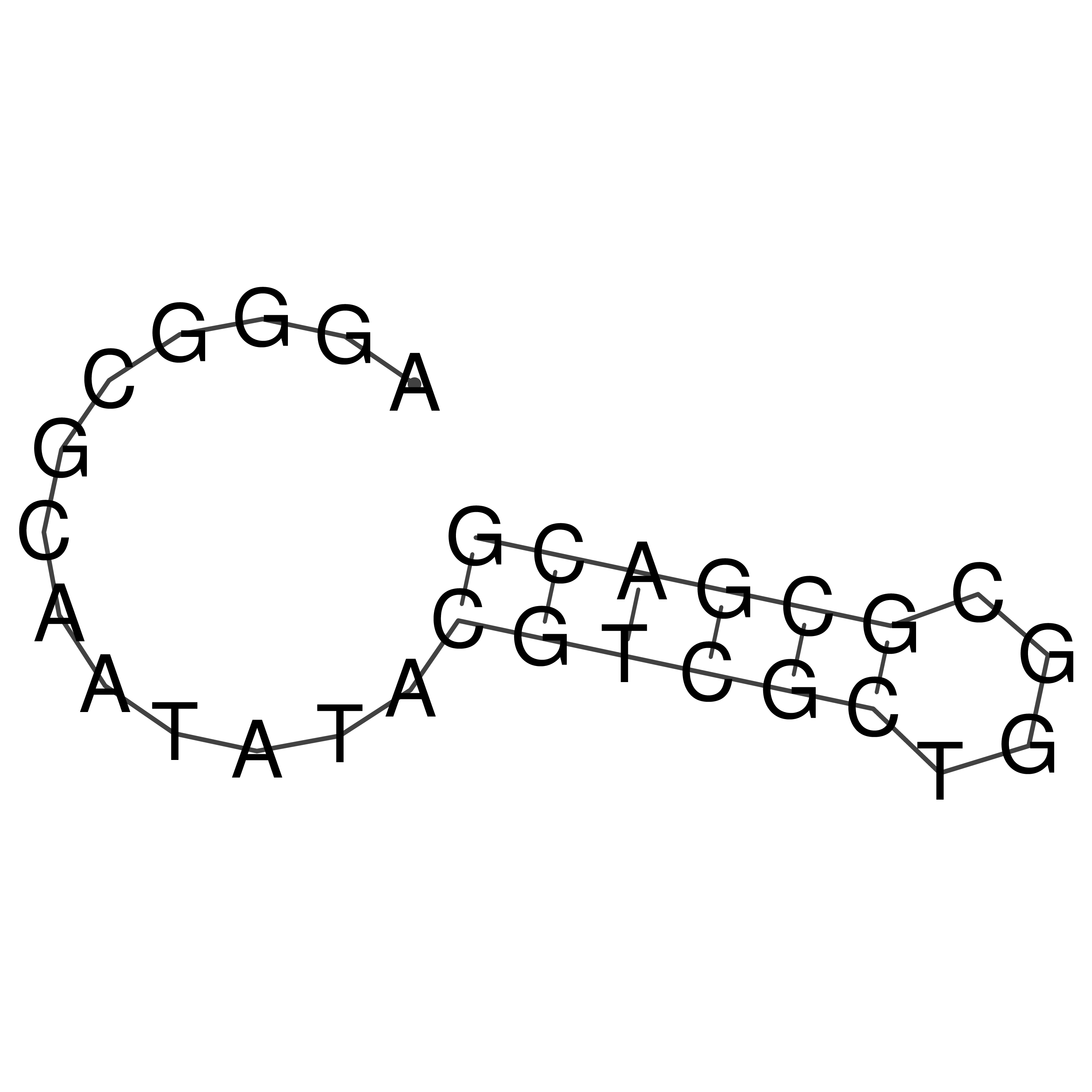
Relaxase
| ID | 2859 | GenBank | WP_095926565 |
| Name | traA_BWO76_RS10325_B401|pSymB |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169946.70 Da Isoelectric Point: 9.5404
>WP_095926565.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAAIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDDRFGSNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSQKIVSERRQRAINQPRGIRM
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
EALRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAAIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDDRFGSNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRIQSSQKIVSERRQRAINQPRGIRM
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2691 | GenBank | WP_014530096 |
| Name | tcpA_BWO76_RS06640_B401|pSymB |
UniProt ID | _ |
| Length | 946 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 946 a.a. Molecular weight: 101605.36 Da Isoelectric Point: 4.5919
>WP_014530096.1 FtsK/SpoIIIE domain-containing protein [Sinorhizobium meliloti]
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEES
RIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQADARLAYLSDFAFFEFMPL
EVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEPV
TAAPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAIVPASEPAPEAPVTRAAITMPAVIQRSSPSLPPI
GAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEF
EPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLAL
CLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIP
HLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTGE
PLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKA
NFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPE
YLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVE
RMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
MRIPRTNFSPAALYDANHADDYEDPHPAAQGAAHVPAAPEAAAHDHLFEAPEAPRRAPGHRDEETGYAGR
AARLYGHGSAYAPAEVTVSTRDPLPTLAEITGLSGEPGWESHFFLSPNVRFTRTPERELMKRHPPAPEES
RIEADEAAAEASDAETVMDVVPAEPAPSVVETELPSYSPSELLRVLTQQLPSWSAARSQAPEASVTKPAI
TESVAVAEEGPATSETTALPVTDEVPVVPHALPVANLAPESAAVEEDVPHQADARLAYLSDFAFFEFMPL
EVAAVPPTVTEPVKEAARIPAPIAAAPKVSPPKIVAAMPVEIRPQPPTAISSLFRVVECRRPEPATAEPV
TAAPQDIAAEPAAAEAAALPAHQFVETPAPALAEPAIVPASEPAPEAPVTRAAITMPAVIQRSSPSLPPI
GAIEPLQGGDAYEFPSKELLQEPPQGQGFFMTQEQLEQNAGLLESVLEDFGVKGEIIHVRPGPVVTLYEF
EPAPGVKSSRVIGLADDIARSMSALSARVAVVPGRNVIGIELPNATRETVYFRELIESGDFQKTGCKLAL
CLGKTIGGEPVIAELAKMPHLLVAGTTGSGKSVAINTMILSLLYRLKPEECRLIMVDPKMLELSVYDGIP
HLLTPVVTDPKKAVMALKWAVREMEDRYRKMSRLGVRNIDGYNQRAAAAREKGAPILATVQTGFEKGTGE
PLFEQQEMDLSPMPYIVVIVDEMADLMMVAGKEIEGAIQRLAQMARAAGIHLIMATQRPSVDVITGTIKA
NFPTRISFQVTSKIDSRTILGEQGAEQLLGQGDMLHMAGGGRIARVHGPFVSDQEVEHVVAHLKTQGRPE
YLETVTADEEEEEVEEDQGAVFDKSAIAAEDGNELYDQAVKVVLRDKKCSTSYIQRRLGIGYNRAASLVE
RMEKDGLVGPANHVGKREIIYGNRDNAPKPESDDLD
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
Host bacterium
| ID | 4289 | GenBank | NZ_CP019484 |
| Plasmid name | B401|pSymB | Incompatibility group | - |
| Plasmid size | 1584717 bp | Coordinate of oriT [Strand] | 867761..867789 [+] |
| Host baterium | Sinorhizobium meliloti strain B401 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | actP |
| Degradation gene | - |
| Symbiosis gene | - |
| Anti-CRISPR | AcrIIA7 |