Detailed information of oriT
oriT
The information of the oriT region
| oriTDB ID | 103848 |
| Name | oriT_B401|pSymA |
| Organism | Sinorhizobium meliloti strain B401 |
| Sequence Completeness | - |
| NCBI accession of oriT (coordinates [strand]) | NZ_CP019483 (479650..479678 [+], 29 nt) |
| oriT length | 29 nt |
| IRs (inverted repeats) | 14..19, 24..29 (CGTCGC..GCGACG) |
| Location of nic site | _ |
| Conserved sequence flanking the nic site |
_ |
| Note | Predicted by oriTfinder 2.0 |
oriT sequence
Download Length: 29 nt
>oriT_B401|pSymA
AGGGCGCAATATACGTCGCTGGCGCGACG
AGGGCGCAATATACGTCGCTGGCGCGACG
Visualization of oriT structure
oriT secondary structure
Predicted by RNAfold.
Download structure file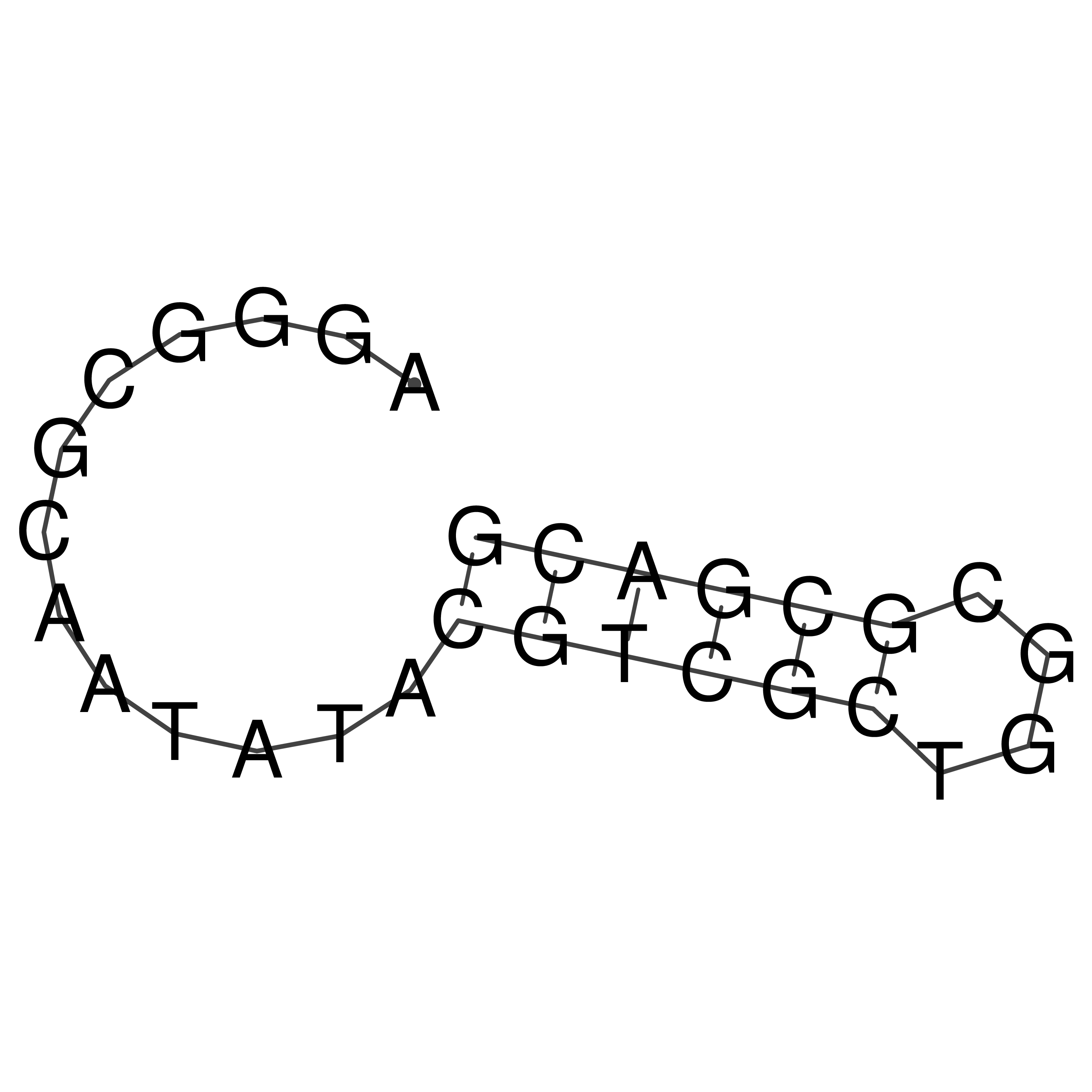
Relaxase
| ID | 2858 | GenBank | WP_095926488 |
| Name | traA_BWO76_RS02715_B401|pSymA |
UniProt ID | _ |
| Length | 1539 a.a. | PDB ID | |
| Note | Predicted by oriTfinder 2.0 | ||
Relaxase protein sequence
Download Length: 1539 a.a. Molecular weight: 169991.68 Da Isoelectric Point: 9.5432
>WP_095926488.1 Ti-type conjugative transfer relaxase TraA [Sinorhizobium meliloti]
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
TSLRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAAIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDDRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRVQSSQEIISERQRRVIDRARGVTR
MAIMFVRAQVIGRGAGRSIVSAAAYRHRTRMIDEQAGTSFSYRGGASELVHEELALPDDIPAWLKAAIDG
RSVAKASEALWNAVEAHETRADAQLARELIIALPEELTRAENIALVREFVRDNLTSKGMVADWVYHDKDG
NPHIHLMTALRPLTEEGFGPKKVPVLGEDGEPLRVVTPDRPNGKIVYKLWAGDKETIKAWKIAWAETANR
HLALAGHEIRLDGRSYAEQGLDGIAQKHLGPEKAALARKGIAMYFAPADLARRQEMADRLLAEPGLLLKQ
LGNERSTFDERDIAKALHRYVDDPVDFANIRARLMASDDLVLLKPQQVDAETGKAKQPAVFTTREMLRLE
YAMARSAEVLSRRKGFGVSNARAAAAVRSIETADTEKPFRLDPEQVDAVRHVTRDNAIAAVVGLAGAGKS
TLLAAARVAWEGEGRRVIGAALAGKAAEGLEDSSGIRSRTLASWELAWESGRKQLNRGDVLVIDEAGMVS
SQQMARILKAVEDAGAKAVLVGDAMQLQPIEAGAAFRAITERIGFAELAGVRRQRDAWARDASRLFARSK
VEEGLDAYAQQGRIVETETRAEIVDRIVADWANARRDLLQKSADGEHPGRLRGDELLVLAHTNDDVRKLN
TSLRNVMIGEGALAGAREFQTARGLREFAAGDRIIFLENARFVEPRARRLGPQYVKNGMLGTVVSTGDRR
GDTLLSVRLDSGRDVVISEDSYRNVDHGYAATIHKSQGSTVDRTFVLATGMMDQHLTYVAMTRHRDRADL
YAAKEDFEAKPEWGRKPRVDHAAGVTGELVKEGMAKFRPNDEDADESPYADIRTDDGTVHRLWGVSLPKA
LKDAGVAEGDTITLRKDGVERVKVQVPIVDAQTGEKRFEERQVDRNVWSASQLETAAARRERIERESHRP
QLFKQLVERLSRSGAKTTTLDFEDEAGYQAQARDFARRRGLYHLSLVAAGMEAEVLRRWAAIAEKREQVA
KLWERASVALGFAIERERRVAYNEERTETLSTGIPSDGKYLVPPTTTFSRSVAEDARLAQLSSQRWKERE
AIVHPVLAKIYRDPDGALSALNALASDAAIEPRKLAEDLGLAPERLGRLRGSELVVDGRAARDERTAATA
ALSELLPLARAHATEFRRNAERFGIREQQRRAHMALSVPALSKTAMARLVEIEAVREQGGDDAYRTAFAY
AVEDRLLVQEVKAVNEALTARFGWSAFTAKADVIAERNMSERMPEDLAPERREKLTRLFAVIRRFAEEQH
LAERQDRSKIVAGASVELGKETFAVLPMLAAVTEFKTTVDEEARERALAAPHYAHHRAALVETATRVWRD
PADAIGKIEDLIVKGFAGERIAAAVTNDPAAYGALRGSDRIMDKLLAVGRERKGALQAVPEAASRIRSLG
ASYASALDAETRSITEERRRMAVAIPGLSPAAEDALKRLAAQIKNKDGKLDVAAGSLDPRIAREFAKVSR
ALDDRFGRNAILRGETDVINRVSPAQRRAFEAMRDRLQVLQQAVRVQSSQEIISERQRRVIDRARGVTR
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4CP
| ID | 2690 | GenBank | WP_014990014 |
| Name | traG_BWO76_RS02700_B401|pSymA |
UniProt ID | _ |
| Length | 639 a.a. | PDB ID | _ |
| Note | Predicted by oriTfinder 2.0 | ||
T4CP protein sequence
Download Length: 639 a.a. Molecular weight: 70274.26 Da Isoelectric Point: 9.4585
>WP_014990014.1 Ti-type conjugative transfer system protein TraG [Sinorhizobium meliloti]
MALKAKPHPSLLVILFPVAVTTAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
MALKAKPHPSLLVILFPVAVTTAAVYVVGWRWPGLAAGMSGKTAYWFLRAAPVPALLFGPLAGLLAVWAL
PLHRRRPVAMASLACFLTVAGFYALREFGRLSPSVESGALSWDRALSYLDMVAVVGAVVGFMAVAVSARI
STVVPEPVKRAKRGTFGDADWLPMAAAGKLFPPDGEIVIGERYRVDKDIVHELPFEPNDPATWGQGGKAL
LLTYRQDFDSTHMLFFAGSGGYKTTSNVVPTALRYTGPLICLDPSTEVAPMVIEHRTRVLGREVMVLDPT
NPIMGFNVLDGIEHSRQKEEDIVGIAHMLLSESVRFESSTGSYFQNQAHNLLTGLLAHVMLSPDYAGRRT
LRSLRQIVSEPEPSVLAMLRDIQEHSASAFIRETLGVFTNMTEQTFSGVYSTASKDTQWLSLDSYAALVC
GNAFKSSDIVSGKKDVFLNIPASILRSYPGIGRVIIGSLINAMIQADGSFKRRALFMLDEVDLLGYMRLL
EEARDRGRKYGISMMLLYQSLGQLERHFGRDGAVSWIDGCAFASYAAVKALDTARNISAQCGEMTVEVKG
SSRNIGWDTKNSASRKSESVNYQRRPLIMPHEITQSMRKDEQIIIVQGHSPIRCGRAIYFRRKDMNEAAK
ANRFVKAIP
Protein domains
Predicted by InterproScan.
Protein structure
No available structure.
T4SS
T4SS were predicted by using oriTfinder2.
Region 1: 589217..598505
| Locus tag | Coordinates | Strand | Size (bp) | Protein ID | Product | Description |
|---|---|---|---|---|---|---|
| BWO76_RS03305 (BWO76_03255) | 584833..585855 | - | 1023 | WP_095918936 | alcohol dehydrogenase AdhP | - |
| BWO76_RS03310 (BWO76_03260) | 586103..586782 | - | 680 | Protein_605 | YoaK family protein | - |
| BWO76_RS03315 (BWO76_03265) | 587034..587645 | + | 612 | WP_095918937 | TetR/AcrR family transcriptional regulator | - |
| BWO76_RS03320 (BWO76_03270) | 587680..588906 | + | 1227 | WP_095918939 | MFS transporter | - |
| BWO76_RS03325 (BWO76_03275) | 589217..590206 | - | 990 | WP_095918940 | P-type DNA transfer ATPase VirB11 | virB11 |
| BWO76_RS03330 (BWO76_03280) | 590214..591098 | - | 885 | Protein_609 | TrbI/VirB10 family protein | - |
| BWO76_RS03335 (BWO76_03285) | 591108..591962 | - | 855 | WP_095918942 | P-type conjugative transfer protein VirB9 | virB9 |
| BWO76_RS03340 (BWO76_03290) | 591962..592633 | - | 672 | WP_095918943 | virB8 family protein | virB8 |
| BWO76_RS03345 (BWO76_03295) | 592638..592952 | - | 315 | WP_095918945 | hypothetical protein | - |
| BWO76_RS03350 (BWO76_03300) | 592952..593884 | - | 933 | WP_198389900 | type IV secretion system protein | virB6 |
| BWO76_RS03355 (BWO76_03305) | 593890..594123 | - | 234 | WP_088198224 | EexN family lipoprotein | - |
| BWO76_RS03360 (BWO76_03310) | 594120..594821 | - | 702 | WP_095918948 | P-type DNA transfer protein VirB5 | virB5 |
| BWO76_RS03365 (BWO76_03315) | 594821..597217 | - | 2397 | WP_198389902 | VirB4 family type IV secretion system protein | virb4 |
| BWO76_RS03370 (BWO76_03320) | 597192..597533 | - | 342 | WP_013845463 | type IV secretion system protein VirB3 | virB3 |
| BWO76_RS03375 (BWO76_03325) | 597538..597837 | - | 300 | WP_088196212 | TrbC/VirB2 family protein | virB2 |
| BWO76_RS03380 (BWO76_03330) | 597834..598505 | - | 672 | WP_095918951 | lytic transglycosylase domain-containing protein | virB1 |
| BWO76_RS03385 (BWO76_03335) | 598509..599027 | - | 519 | WP_095918952 | hypothetical protein | - |
| BWO76_RS03390 (BWO76_03340) | 599124..599492 | + | 369 | WP_013845467 | hypothetical protein | - |
| BWO76_RS31525 | 599607..600002 | - | 396 | WP_234827448 | hypothetical protein | - |
| BWO76_RS03400 (BWO76_03350) | 600077..600724 | - | 648 | WP_095918954 | dihydrofolate reductase family protein | - |
| BWO76_RS03405 (BWO76_03355) | 600774..601622 | - | 849 | WP_095918956 | alpha/beta fold hydrolase | - |
Host bacterium
| ID | 4288 | GenBank | NZ_CP019483 |
| Plasmid name | B401|pSymA | Incompatibility group | - |
| Plasmid size | 1096825 bp | Coordinate of oriT [Strand] | 479650..479678 [+] |
| Host baterium | Sinorhizobium meliloti strain B401 |
Cargo genes
| Drug resistance gene | - |
| Virulence gene | htpB |
| Metal resistance gene | nia, ncrA |
| Degradation gene | - |
| Symbiosis gene | nodD, fixN, fixO, fixU, nifB, fixX, fixC, fixB, fixA, nifH, nifD, nifK, nifE, nifX, nodH, nodF, nodE, nodJ, nodI, nodC, nodB, nodA, nifN, nodM, fixS, fixG |
| Anti-CRISPR | - |